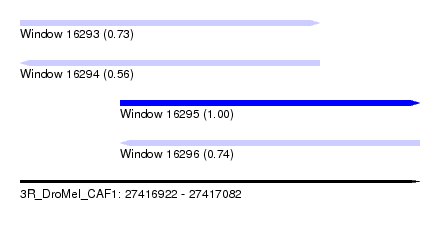
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,416,922 – 27,417,082 |
| Length | 160 |
| Max. P | 0.997473 |

| Location | 27,416,922 – 27,417,042 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -46.16 |
| Consensus MFE | -38.10 |
| Energy contribution | -38.58 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27416922 120 + 27905053 GCUUGGUGCUGUAAUAGGCACCUGAAAGUGCCCAAUCUGCCAGAGGGAGUACCCUUGCCGUACUCCCUCGUGACGGUCCUGGUGGACCGGAUCGACUUCACCAGUGUCUCACCCCAGGGC ((((((((((......))))))...))))((((.........((((((((((.......))))))))))((((((((((....))))))....(((.........)))))))....)))) ( -52.80) >DroSec_CAF1 19020 120 + 1 GCUUGGUGCUGUAAUAGGCACCUCAAAGUGCCCAAUCUGCCGGAGGGAGUGCCCUUGCCAUACUCCAUCGUGACGGUCCUGGUGGACCGGAUCGACUUCACCAGUGUCUCACCCCAGGGC ((((((((((......))))))...))))((((........(((((((((((....))...))))).(((...((((((....))))))...)))))))....(((...)))....)))) ( -43.00) >DroSim_CAF1 18843 120 + 1 GCUUGGUGCUGUAAUAGACACCUCAAAGUGCCCAAUCUGCCGGAGGGAGUGCCCUUGCCGUACUCCCUUGUGACGGUCCUGGUGGACCGGAUCGACUUCACCAGUGUCUCACCCCAGGGC (((((((((((...))).))))...))))((((.........((((((((((.......))))))))))((((((((((....))))))....(((.........)))))))....)))) ( -45.70) >DroEre_CAF1 19393 120 + 1 GCUUGGUGCUGUCAUAGGCACCUGGUAGUCCCCAAUCUGCCGGAGGGAGUACCUCUACCGUACUCCAUCGUGACGGUCCUGGUGGACCGAAUCGACUUCACCAGUGUCGCACCCCUAGGC (((((((((.(.(((.((.....(((((........)))))(((((((((((.......))))))).(((...((((((....))))))...))))))).)).))).))))))...)))) ( -49.30) >DroYak_CAF1 19260 120 + 1 GCUUGGUGCUGUCACAGGCACCUGAAAGUGCCCAAUCUGCCGGAGGGAGUCCCCUUACCGUACUCCAUCGUGACGGUCCUGGUGGACCGAAUUGACUUCACUAGUGUCUCACCCAAUGGC (((.((((..(.(((.(((((......))))).........((((((((((........).))))).(((...((((((....))))))...)))))))....))).).))))....))) ( -40.00) >consensus GCUUGGUGCUGUAAUAGGCACCUGAAAGUGCCCAAUCUGCCGGAGGGAGUACCCUUGCCGUACUCCAUCGUGACGGUCCUGGUGGACCGGAUCGACUUCACCAGUGUCUCACCCCAGGGC (((.((((((......))))))....))).........(((.((.(((((((.......))))))).))((((((((((....))))))....(((.........))))))).....))) (-38.10 = -38.58 + 0.48)


| Location | 27,416,922 – 27,417,042 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -47.62 |
| Consensus MFE | -37.94 |
| Energy contribution | -38.14 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27416922 120 - 27905053 GCCCUGGGGUGAGACACUGGUGAAGUCGAUCCGGUCCACCAGGACCGUCACGAGGGAGUACGGCAAGGGUACUCCCUCUGGCAGAUUGGGCACUUUCAGGUGCCUAUUACAGCACCAAGC (((....))).......(((((..(((((((.(((((....)))))((((.((((((((((.......)))))))))))))).)))))((((((....))))))....))..)))))... ( -56.20) >DroSec_CAF1 19020 120 - 1 GCCCUGGGGUGAGACACUGGUGAAGUCGAUCCGGUCCACCAGGACCGUCACGAUGGAGUAUGGCAAGGGCACUCCCUCCGGCAGAUUGGGCACUUUGAGGUGCCUAUUACAGCACCAAGC (((....))).......(((((..((((...((((((....))))))....((.(((((...((....))))))).))))))....((((((((....))))))))......)))))... ( -45.00) >DroSim_CAF1 18843 120 - 1 GCCCUGGGGUGAGACACUGGUGAAGUCGAUCCGGUCCACCAGGACCGUCACAAGGGAGUACGGCAAGGGCACUCCCUCCGGCAGAUUGGGCACUUUGAGGUGUCUAUUACAGCACCAAGC (((....))).......(((((..((((...((((((....)))))).....(((((((...((....))))))))).))))....((((((((....))))))))......)))))... ( -45.20) >DroEre_CAF1 19393 120 - 1 GCCUAGGGGUGCGACACUGGUGAAGUCGAUUCGGUCCACCAGGACCGUCACGAUGGAGUACGGUAGAGGUACUCCCUCCGGCAGAUUGGGGACUACCAGGUGCCUAUGACAGCACCAAGC .......(((((......((.(..((((...((((((....))))))...))))(((((((.......)))))))).))((((..((((......)))).)))).......))))).... ( -48.00) >DroYak_CAF1 19260 120 - 1 GCCAUUGGGUGAGACACUAGUGAAGUCAAUUCGGUCCACCAGGACCGUCACGAUGGAGUACGGUAAGGGGACUCCCUCCGGCAGAUUGGGCACUUUCAGGUGCCUGUGACAGCACCAAGC .......((((.(((.........))).....(((((....)))))(((((......((.(((..((((....))))))))).....(((((((....))))))))))))..)))).... ( -43.70) >consensus GCCCUGGGGUGAGACACUGGUGAAGUCGAUCCGGUCCACCAGGACCGUCACGAUGGAGUACGGCAAGGGCACUCCCUCCGGCAGAUUGGGCACUUUCAGGUGCCUAUUACAGCACCAAGC (((....))).......(((((..((((...((((((....))))))....((.(((((.(.......).))))).))))))....((((((((....))))))))......)))))... (-37.94 = -38.14 + 0.20)

| Location | 27,416,962 – 27,417,082 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -52.24 |
| Consensus MFE | -44.94 |
| Energy contribution | -45.30 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27416962 120 + 27905053 CAGAGGGAGUACCCUUGCCGUACUCCCUCGUGACGGUCCUGGUGGACCGGAUCGACUUCACCAGUGUCUCACCCCAGGGCACCCAAGACAUCCGUCUGUCUGGCCACGUGUCCUGGGUGG ..((((((((((.......))))))))))((((((((((....))))))....(((.........)))))))(((((((((((((.((((......)))))))....))))))))))... ( -60.00) >DroSec_CAF1 19060 120 + 1 CGGAGGGAGUGCCCUUGCCAUACUCCAUCGUGACGGUCCUGGUGGACCGGAUCGACUUCACCAGUGUCUCACCCCAGGGCACCCAAGACAUCCGCCUGUCUGGCCAUGUGUCGUGGGUGG .(((((((((((....))...))))).(((...((((((....))))))...)))))))............((....))((((((.(((((..(((.....)))...))))).)))))). ( -48.70) >DroSim_CAF1 18883 120 + 1 CGGAGGGAGUGCCCUUGCCGUACUCCCUUGUGACGGUCCUGGUGGACCGGAUCGACUUCACCAGUGUCUCACCCCAGGGCACCCAAGACAUCCGCCUGUCUGGCCAUGUGUCCUGGGUGG ..((((((((((.......))))))))))((((((((((....))))))....(((.........)))))))(((((((((((((.((((......)))))))....))))))))))... ( -57.30) >DroEre_CAF1 19433 120 + 1 CGGAGGGAGUACCUCUACCGUACUCCAUCGUGACGGUCCUGGUGGACCGAAUCGACUUCACCAGUGUCGCACCCCUAGGCACCCAGGACAUCCGCCUAUCCGGCCAUGUCUCCUGGGUGG .((..(((((((.......)))))))...(((.(((..((((((((..........))))))))..)))))).))....(((((((((.((..(((.....)))...)).))))))))). ( -51.00) >DroYak_CAF1 19300 120 + 1 CGGAGGGAGUCCCCUUACCGUACUCCAUCGUGACGGUCCUGGUGGACCGAAUUGACUUCACUAGUGUCUCACCCAAUGGCACCCAAGACAUCCGCCUAUCUGGCCACGUCUCCUGGGUGG (((((((....))))..)))....((((.((((((((((....))))))....(((.........)))))))...))))((((((((((....(((.....)))...))))..)))))). ( -44.20) >consensus CGGAGGGAGUACCCUUGCCGUACUCCAUCGUGACGGUCCUGGUGGACCGGAUCGACUUCACCAGUGUCUCACCCCAGGGCACCCAAGACAUCCGCCUGUCUGGCCAUGUGUCCUGGGUGG .(((((((((((.......))))))).(((...((((((....))))))...)))))))............((....))((((((.(((((..(((.....)))...))))).)))))). (-44.94 = -45.30 + 0.36)

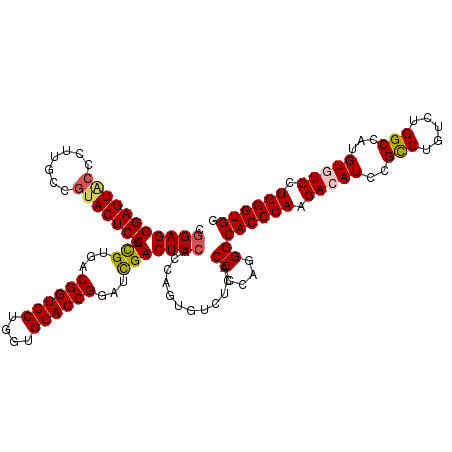

| Location | 27,416,962 – 27,417,082 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -50.90 |
| Consensus MFE | -42.78 |
| Energy contribution | -43.82 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27416962 120 - 27905053 CCACCCAGGACACGUGGCCAGACAGACGGAUGUCUUGGGUGCCCUGGGGUGAGACACUGGUGAAGUCGAUCCGGUCCACCAGGACCGUCACGAGGGAGUACGGCAAGGGUACUCCCUCUG ...((((((.(((....(((((((......)))).)))))).))))))(((((((.........)))....((((((....))))))))))((((((((((.......)))))))))).. ( -57.10) >DroSec_CAF1 19060 120 - 1 CCACCCACGACACAUGGCCAGACAGGCGGAUGUCUUGGGUGCCCUGGGGUGAGACACUGGUGAAGUCGAUCCGGUCCACCAGGACCGUCACGAUGGAGUAUGGCAAGGGCACUCCCUCCG .((((((.((((..(.(((.....))).).)))).))))))....((((...............((((...((((((....))))))...))))(((((...((....))))))))))). ( -44.70) >DroSim_CAF1 18883 120 - 1 CCACCCAGGACACAUGGCCAGACAGGCGGAUGUCUUGGGUGCCCUGGGGUGAGACACUGGUGAAGUCGAUCCGGUCCACCAGGACCGUCACAAGGGAGUACGGCAAGGGCACUCCCUCCG .(((((((((((..(.(((.....))).).)))))))))))((((...(((((((.........)))....((((((....)))))))))).))))....(((..((((....))))))) ( -53.00) >DroEre_CAF1 19433 120 - 1 CCACCCAGGAGACAUGGCCGGAUAGGCGGAUGUCCUGGGUGCCUAGGGGUGCGACACUGGUGAAGUCGAUUCGGUCCACCAGGACCGUCACGAUGGAGUACGGUAGAGGUACUCCCUCCG .(((((((((.(..(.(((.....))).).).)))))))))....((((...............((((...((((((....))))))...))))(((((((.......))))))))))). ( -52.60) >DroYak_CAF1 19300 120 - 1 CCACCCAGGAGACGUGGCCAGAUAGGCGGAUGUCUUGGGUGCCAUUGGGUGAGACACUAGUGAAGUCAAUUCGGUCCACCAGGACCGUCACGAUGGAGUACGGUAAGGGGACUCCCUCCG .(((((((((.(..(.(((.....))).).).)))))))))(((((..(((((((.........)))....((((((....)))))))))))))))....(((..((((....))))))) ( -47.10) >consensus CCACCCAGGACACAUGGCCAGACAGGCGGAUGUCUUGGGUGCCCUGGGGUGAGACACUGGUGAAGUCGAUCCGGUCCACCAGGACCGUCACGAUGGAGUACGGCAAGGGCACUCCCUCCG .(((((((((((..(.(((.....))).).)))))))))))((((...(((((((.........)))....((((((....)))))))))).))))....(((..((((....))))))) (-42.78 = -43.82 + 1.04)

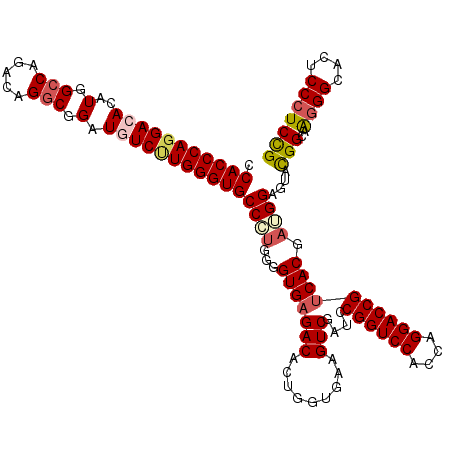
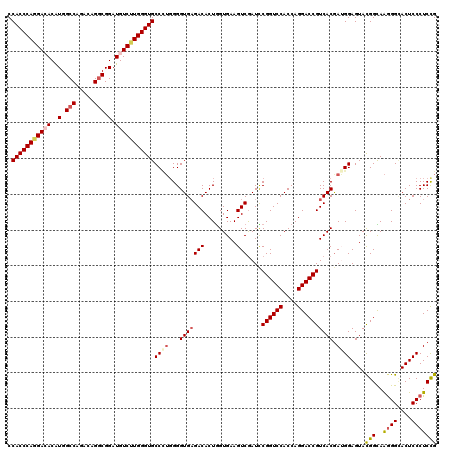
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:52:02 2006