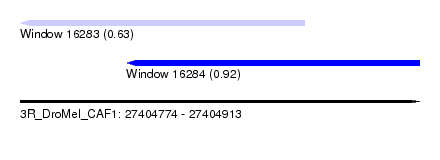
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,404,774 – 27,404,913 |
| Length | 139 |
| Max. P | 0.921050 |

| Location | 27,404,774 – 27,404,873 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 85.72 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -26.82 |
| Energy contribution | -27.73 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27404774 99 - 27905053 CCAUAUGCACCCGCAUCAUCCAGGCGGACCUAUGGGUCACCCACAUGGCCCACAUCCGCACAUGGGUGGUCCACCUCCAAUGCGAGGAAUGAGCCCCAU ......((....(.((((((((.(((((....(((((((......)))))))..)))))...)))))))).).((((......)))).....))..... ( -36.00) >DroSec_CAF1 6900 99 - 1 CCAUAUGCACCCGCAUCAUCCAGGCGGACCCAUGGGUCACCCACAUGGCCCACAUCCGCACAUGAGUGGUCCGCCUCCAAUGCGAGGCAUGAGCCCCAU ...(((((.(.(((((.....((((((((((.(((((((......)))))))(((......))).).)))))))))...))))).))))))........ ( -40.20) >DroEre_CAF1 7156 99 - 1 CCAUAUGCACCCGCAUCAUCCAGGCGGACCCAUGGGUCACCCACAUGGUCCACAUCCGCACAUGGGUGGUCCGCCUCCGAUGCGUGGCAUGAGCCCCAU ...(((((.(.((((((....(((((((((((((((....)).)))))....((((((....))))))))))))))..)))))).))))))........ ( -43.90) >DroWil_CAF1 6802 93 - 1 UCACAUGCAUCCACAUCAUCCAGGCGGUCCGAUGGGACAUCCACAUGGACCACAU---CAUAUGGGUGGACCG---CCCAUGCGCGGCAUGAGUCCAAU ...(((((.(((((.((((....(.(((((.(((((....)).)))))))).)..---...))))))))).((---(......))))))))........ ( -33.50) >DroYak_CAF1 6456 99 - 1 CCAUAUGCACCCGCAUCAUCCAGGCGGACCCAUGGGUCAUCCACAUGGUCCCCAUCCGCACAUGGGUGGUCCGCCUCCAAUGCGAGGCAUGAGCCCCAU ...(((((.(.(((((.....(((((((((((((((....)).))))...(((((......))))).)))))))))...))))).))))))........ ( -41.30) >DroAna_CAF1 6773 99 - 1 GCACAUGCAUCCCCAUCAUCCAGGUGGUCCAAUGGGUCAUCCCCACGGUCCCCAUCCGCACAUGGGAGGGCCUCCGCCAAUGCGAGGAAUGAGUCCGAU ((....))(((....((((((.((.(((((..((((.....)))).....(((((......))))).))))).))((....))..)).))))....))) ( -31.80) >consensus CCAUAUGCACCCGCAUCAUCCAGGCGGACCCAUGGGUCACCCACAUGGUCCACAUCCGCACAUGGGUGGUCCGCCUCCAAUGCGAGGCAUGAGCCCCAU ...(((((...(((((.....(((((((((..((((...))))((((((........)).))))...)))))))))...)))))..)))))........ (-26.82 = -27.73 + 0.92)



| Location | 27,404,811 – 27,404,913 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.92 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -25.32 |
| Energy contribution | -26.35 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27404811 102 - 27905053 GCAUGGGCGGUGGACCUCCUCCCCCGCCUCACAUGCAUCCCCAUAUGCACCCGCAUCAUCCAGGCGGACCUAUGGGUCACCCACAUGGCCCACAUCCGCACA ((((((((((.(((.....))).)))))...)))))........((((....)))).......(((((....(((((((......)))))))..)))))... ( -40.00) >DroPse_CAF1 7506 93 - 1 GCAUGGGCGGUGGG---------CCGCCGCAUAUGCAUCCCCACAUGCAUCCGCACCAUCCGGGCGGACCGAUGGGACAUCCCCAUGGACCCCCGCAUCACA ((((((((((....---------)))))((....)).......))))).(((((.((....)))))))((.(((((.....))))))).............. ( -39.30) >DroSec_CAF1 6937 102 - 1 GCAUGGGCGGUGGACCUCCUCCCCCGCCGCACAUGCAUCCCCAUAUGCACCCGCAUCAUCCAGGCGGACCCAUGGGUCACCCACAUGGCCCACAUCCGCACA (((((.(((((((..........))))))).)))))........((((....)))).......(((((....(((((((......)))))))..)))))... ( -41.90) >DroYak_CAF1 6493 102 - 1 GCAUGGGUGGUGGACCUCCUCCCCCGCCUCACAUGCAUCCCCAUAUGCACCCGCAUCAUCCAGGCGGACCCAUGGGUCAUCCACAUGGUCCCCAUCCGCACA ((..((((((.(((((........(((((...(((.........((((....)))))))..)))))((((....))))........)))))))))))))... ( -37.60) >DroAna_CAF1 6810 102 - 1 GCAUGGGUGGAGCACCACCGCCGCCGCCGCACAUGCACCCGCACAUGCAUCCCCAUCAUCCAGGUGGUCCAAUGGGUCAUCCCCACGGUCCCCAUCCGCACA ((..((((((.(.(((((((((((....))..(((((........)))))............))))))....((((.....)))).))).)))))))))... ( -33.30) >DroPer_CAF1 6951 93 - 1 GCAUGGGCGGUGGC---------CCGCCGCAUAUGCAUCCCCACAUGCAUCCGCACCAUCCGGGCGGACCGAUGGGUCAUCCCCAUGGACCCCCGCAUCACA (((((((.((((((---------(((.((....(((((......)))))(((((.((....))))))).)).))))))))))))))((....))))...... ( -41.90) >consensus GCAUGGGCGGUGGACCUCCUCCCCCGCCGCACAUGCAUCCCCACAUGCACCCGCAUCAUCCAGGCGGACCCAUGGGUCAUCCACAUGGACCCCAUCCGCACA (((((.(((((((..........))))))).))))).........(((..((((.........)))).((.(((((....)).))))).........))).. (-25.32 = -26.35 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:51:51 2006