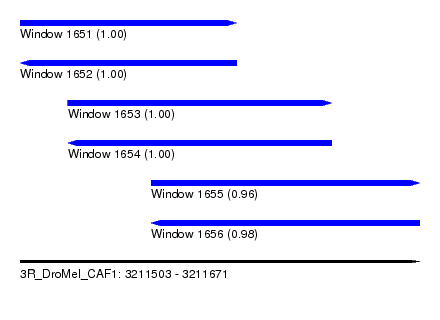
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,211,503 – 3,211,671 |
| Length | 168 |
| Max. P | 0.999942 |

| Location | 3,211,503 – 3,211,594 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 96.28 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -27.86 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.37 |
| SVM RNA-class probability | 0.999882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
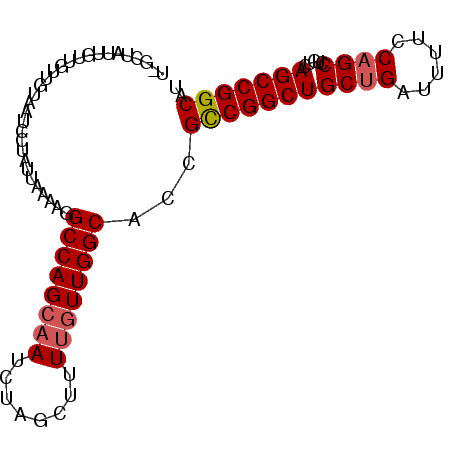
>3R_DroMel_CAF1 3211503 91 + 27905053 U-GCUAUUGUUGUUGUAAUCCUAUUAAAAGGCCAGCAAUCUAGCUUUUGUUGGCACCGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAU .-............................((((((((........))))))))...(((((((((((......)))).....))))))).. ( -31.20) >DroSec_CAF1 36201 91 + 1 U-GCUAUUGUUGUUGUAAUCCUAUUAAAAGGCCAGCAAUCUAGUUUUUAUUGGCACCGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAU (-((((..(((((((....(((......))).)))))))...........)))))..(((((((((((......)))).....))))))).. ( -28.02) >DroSim_CAF1 37694 91 + 1 U-GCUAUUGUUGUUGUAAUCCUAUUAAAAGGCCAGCAAUCUAGCUUUUGUUGGCACCGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAU .-............................((((((((........))))))))...(((((((((((......)))).....))))))).. ( -31.20) >DroEre_CAF1 30132 92 + 1 UGGCUUUUGUUGUUGUAAUCCUAUUAAAAGGCCAGCAAUCUAGCUUUUGUUGGCACCGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAU .((((((((...............))))))((((((((........)))))))).))(((((((((((......)))).....))))))).. ( -33.96) >DroYak_CAF1 19626 90 + 1 U-GCUUUUGUUGUUGUAAUCCUAUUAAAGGGCCAGCUAUCUAGCUUUUGUUGGCACCGUCGGCUGC-GAUUUUCCAGCCUGUCAGCCGGCAU .-..........................((((((((............)))))).))(((((((((-(...........)).)))))))).. ( -26.10) >consensus U_GCUAUUGUUGUUGUAAUCCUAUUAAAAGGCCAGCAAUCUAGCUUUUGUUGGCACCGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAU ..............................((((((((........))))))))...(((((((((((......)))).....))))))).. (-27.86 = -28.30 + 0.44)

| Location | 3,211,503 – 3,211,594 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 96.28 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -23.70 |
| Energy contribution | -23.78 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3211503 91 - 27905053 AUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCGGUGCCAACAAAAGCUAGAUUGCUGGCCUUUUAAUAGGAUUACAACAACAAUAGC-A .(((((((((...(..(....)..)..)))))))))............((((..(((.((.(((......)))....)).)))...))))-. ( -28.70) >DroSec_CAF1 36201 91 - 1 AUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCGGUGCCAAUAAAAACUAGAUUGCUGGCCUUUUAAUAGGAUUACAACAACAAUAGC-A .(((((((((...(..(....)..)..))))))))).(((.........((((....))))(((......)))...............))-) ( -24.40) >DroSim_CAF1 37694 91 - 1 AUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCGGUGCCAACAAAAGCUAGAUUGCUGGCCUUUUAAUAGGAUUACAACAACAAUAGC-A .(((((((((...(..(....)..)..)))))))))............((((..(((.((.(((......)))....)).)))...))))-. ( -28.70) >DroEre_CAF1 30132 92 - 1 AUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCGGUGCCAACAAAAGCUAGAUUGCUGGCCUUUUAAUAGGAUUACAACAACAAAAGCCA ..((((((((...(..(....)..)..))))))))(((((((.(((........))).))))(((.....)))...............))). ( -27.80) >DroYak_CAF1 19626 90 - 1 AUGCCGGCUGACAGGCUGGAAAAUC-GCAGCCGACGGUGCCAACAAAAGCUAGAUAGCUGGCCCUUUAAUAGGAUUACAACAACAAAAGC-A ..((((((((.(.((((((.....)-.)))))......((........))..).))))))))(((.....))).................-. ( -24.50) >consensus AUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCGGUGCCAACAAAAGCUAGAUUGCUGGCCUUUUAAUAGGAUUACAACAACAAUAGC_A ..((((((((...(..(....)..)..))))))))((.((((.(((........))).))))))............................ (-23.70 = -23.78 + 0.08)

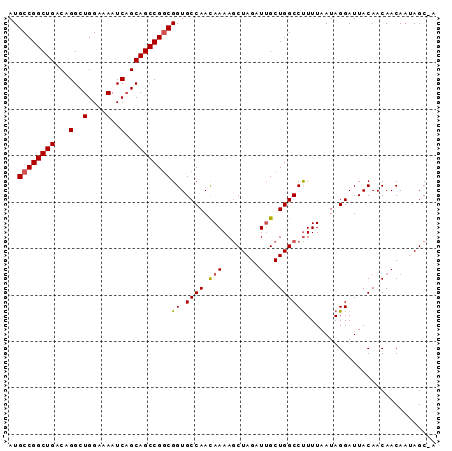
| Location | 3,211,523 – 3,211,634 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 97.12 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -36.70 |
| Energy contribution | -37.34 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
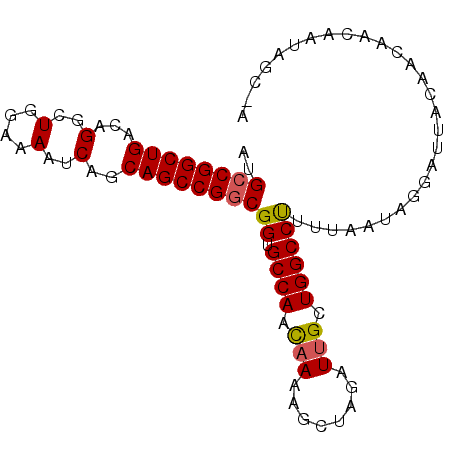
>3R_DroMel_CAF1 3211523 111 + 27905053 UAUUAAAAGGCCAGCAAUCUAGCUUUUGUUGGCACCGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGAUUCGAUUCGGCGAAACAAAG .........((((((((........))))))))...(((((((((((......)))).....))))))).((((((((((..((..........))..)))).)))))).. ( -41.40) >DroSec_CAF1 36221 111 + 1 UAUUAAAAGGCCAGCAAUCUAGUUUUUAUUGGCACCGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGAUUCGAUUCGGCGAAACAAAG .........(((((.((........)).)))))...(((((((((((......)))).....))))))).((((((((((..((..........))..)))).)))))).. ( -35.10) >DroSim_CAF1 37714 111 + 1 UAUUAAAAGGCCAGCAAUCUAGCUUUUGUUGGCACCGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGAUUCGAUUCGGCGAAACAAAG .........((((((((........))))))))...(((((((((((......)))).....))))))).((((((((((..((..........))..)))).)))))).. ( -41.40) >DroEre_CAF1 30153 111 + 1 UAUUAAAAGGCCAGCAAUCUAGCUUUUGUUGGCACCGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGAUUCGAUUCGGCGAAACAAAG .........((((((((........))))))))...(((((((((((......)))).....))))))).((((((((((..((..........))..)))).)))))).. ( -41.40) >DroYak_CAF1 19646 110 + 1 UAUUAAAGGGCCAGCUAUCUAGCUUUUGUUGGCACCGUCGGCUGC-GAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGACUCGUUUCGGCGAAACAAAG .......((((((((............)))))).))(((((((((-(...........)).)))))))).((((((((((...(..........)...)))).)))))).. ( -33.30) >consensus UAUUAAAAGGCCAGCAAUCUAGCUUUUGUUGGCACCGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGAUUCGAUUCGGCGAAACAAAG .........((((((((........))))))))...(((((((((((......)))).....))))))).((((((((((..((..........))..)))).)))))).. (-36.70 = -37.34 + 0.64)

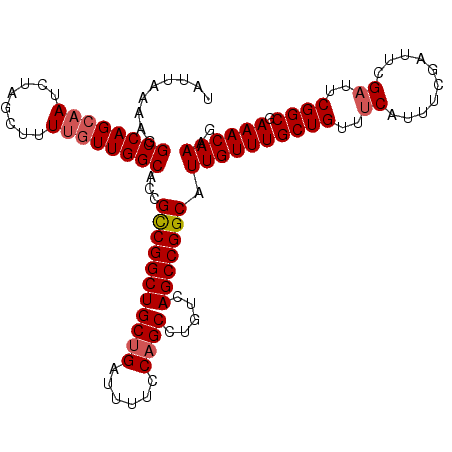
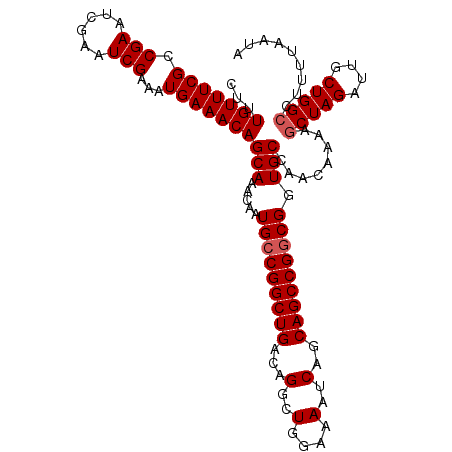
| Location | 3,211,523 – 3,211,634 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 97.12 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -30.88 |
| Energy contribution | -31.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3211523 111 - 27905053 CUUUGUUUCGCCGAAUCGAAUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCGGUGCCAACAAAAGCUAGAUUGCUGGCCUUUUAAUA ...(((((((.(((......)))...)))))))(((.....(((((((((...(..(....)..)..))))))))).)))....(((((((((....)))).))))).... ( -34.10) >DroSec_CAF1 36221 111 - 1 CUUUGUUUCGCCGAAUCGAAUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCGGUGCCAAUAAAAACUAGAUUGCUGGCCUUUUAAUA .((..(((((.((...))...)))))..)).((((((.(...((((((((...(..(....)..)..))))))))(((..........))).).))))))........... ( -31.30) >DroSim_CAF1 37714 111 - 1 CUUUGUUUCGCCGAAUCGAAUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCGGUGCCAACAAAAGCUAGAUUGCUGGCCUUUUAAUA ...(((((((.(((......)))...)))))))(((.....(((((((((...(..(....)..)..))))))))).)))....(((((((((....)))).))))).... ( -34.10) >DroEre_CAF1 30153 111 - 1 CUUUGUUUCGCCGAAUCGAAUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCGGUGCCAACAAAAGCUAGAUUGCUGGCCUUUUAAUA ...(((((((.(((......)))...)))))))(((.....(((((((((...(..(....)..)..))))))))).)))....(((((((((....)))).))))).... ( -34.10) >DroYak_CAF1 19646 110 - 1 CUUUGUUUCGCCGAAACGAGUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUC-GCAGCCGACGGUGCCAACAAAAGCUAGAUAGCUGGCCCUUUAAUA .((((((.(((((....................(((.....)))((((((...(..(....)..)-.)))))).)))))..)))))).(((((....)))))......... ( -29.80) >consensus CUUUGUUUCGCCGAAUCGAAUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCGGUGCCAACAAAAGCUAGAUUGCUGGCCUUUUAAUA ...(((((((.(((......)))...)))))))(((.....(((((((((...(..(....)..)..))))))))).)))........(((((....)))))......... (-30.88 = -31.28 + 0.40)

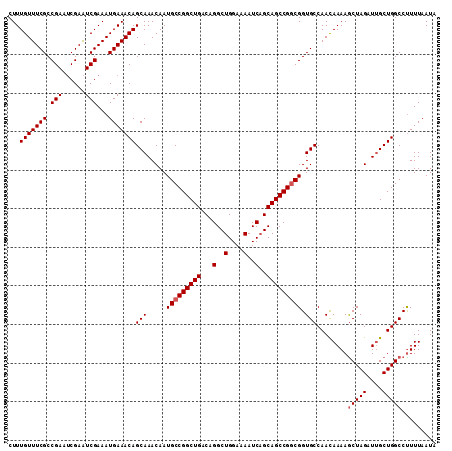
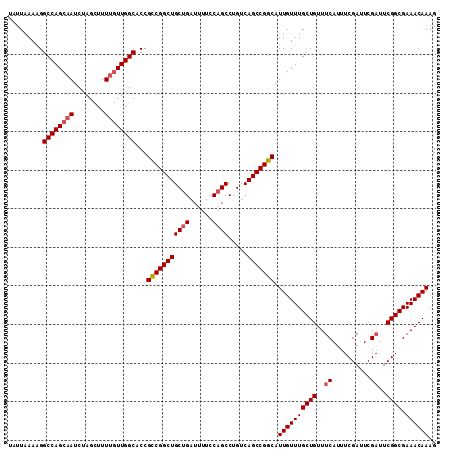
| Location | 3,211,558 – 3,211,671 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 97.35 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.74 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3211558 113 + 27905053 CGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGAUUCGAUUCGGCGAAACAAAGACUUAAAGUGCUCAUUCACCUGAUUUUGCUGGUUUUA .(((((((((((......)))).....)))))))..(((((((((..((..........))..)))).)))))((((((......((....((....))....)).)))))). ( -32.30) >DroSec_CAF1 36256 113 + 1 CGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGAUUCGAUUCGGCGAAACAAAGACUUAAAGUGCUCAUUCACCUGAUUUUGCUGGUUUUA .(((((((((((......)))).....)))))))..(((((((((..((..........))..)))).)))))((((((......((....((....))....)).)))))). ( -32.30) >DroSim_CAF1 37749 113 + 1 CGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGAUUCGAUUCGGCGAAACAAAGACUUAAAGUGCUCAUUCACCUGAUUUUGCUGGUUUUA .(((((((((((......)))).....)))))))..(((((((((..((..........))..)))).)))))((((((......((....((....))....)).)))))). ( -32.30) >DroEre_CAF1 30188 113 + 1 CGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGAUUCGAUUCGGCGAAACAAAGACUUAAAGUGCUCAUUCACCUGAUUUUACUGGUUUUA .(((((((((((......)))).....)))))))..(((((((((..((..........))..)))).)))))((((((...((((.(((......)))...)))))))))). ( -32.30) >DroYak_CAF1 19681 112 + 1 CGUCGGCUGC-GAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGACUCGUUUCGGCGAAACAAAGACUUAAAGUACUCAUUCACCUGCUUUUACUGGUUUUA .(((((((((-(...........)).)))))))).........((((((...((((......)))).))))))((((((.((((((..........))))))....)))))). ( -26.20) >consensus CGCCGGCUGCUGAUUUUCCAGCCUGUCAGCCGGCAUUGUUUGCUGUUUCAUUUCGAUUCGAUUCGGCGAAACAAAGACUUAAAGUGCUCAUUCACCUGAUUUUGCUGGUUUUA .(((((((((((......)))).....)))))))...((((..((((((...((((......)))).)))))).))))...............(((..........))).... (-28.70 = -28.74 + 0.04)

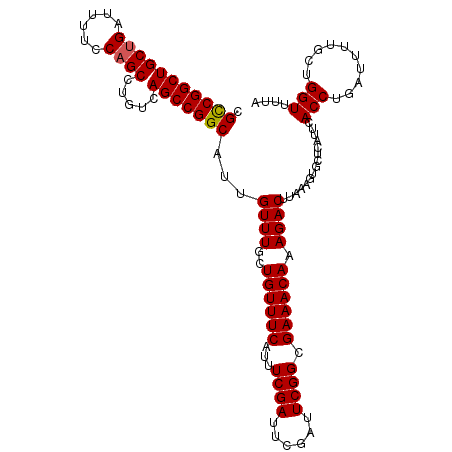
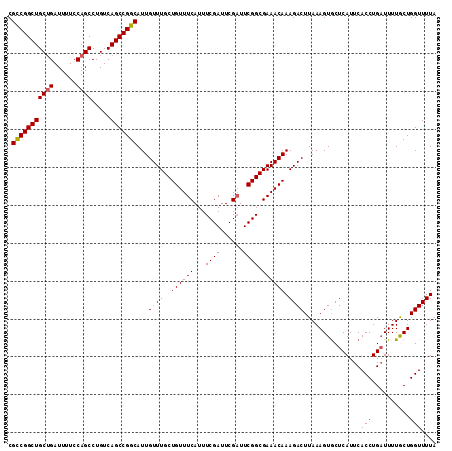
| Location | 3,211,558 – 3,211,671 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 97.35 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -28.28 |
| Energy contribution | -28.24 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3211558 113 - 27905053 UAAAACCAGCAAAAUCAGGUGAAUGAGCACUUUAAGUCUUUGUUUCGCCGAAUCGAAUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCG ........((....(((((((((.(((.((.....)))))...))))))...(((...)))..)))....)).......((((((((...(..(....)..)..)))))))). ( -31.10) >DroSec_CAF1 36256 113 - 1 UAAAACCAGCAAAAUCAGGUGAAUGAGCACUUUAAGUCUUUGUUUCGCCGAAUCGAAUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCG ........((....(((((((((.(((.((.....)))))...))))))...(((...)))..)))....)).......((((((((...(..(....)..)..)))))))). ( -31.10) >DroSim_CAF1 37749 113 - 1 UAAAACCAGCAAAAUCAGGUGAAUGAGCACUUUAAGUCUUUGUUUCGCCGAAUCGAAUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCG ........((....(((((((((.(((.((.....)))))...))))))...(((...)))..)))....)).......((((((((...(..(....)..)..)))))))). ( -31.10) >DroEre_CAF1 30188 113 - 1 UAAAACCAGUAAAAUCAGGUGAAUGAGCACUUUAAGUCUUUGUUUCGCCGAAUCGAAUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCG ........((....(((((((((.(((.((.....)))))...))))))...(((...)))..)))....)).......((((((((...(..(....)..)..)))))))). ( -29.50) >DroYak_CAF1 19681 112 - 1 UAAAACCAGUAAAAGCAGGUGAAUGAGUACUUUAAGUCUUUGUUUCGCCGAAACGAGUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUC-GCAGCCGACG .................((((((.(((.((.....)))))...)))))).......(((.........((((...........))))...((((((.....)-.)))))))). ( -26.70) >consensus UAAAACCAGCAAAAUCAGGUGAAUGAGCACUUUAAGUCUUUGUUUCGCCGAAUCGAAUCGAAAUGAAACAGCAAACAAUGCCGGCUGACAGGCUGGAAAAUCAGCAGCCGGCG ........((.......((((((.(((.((.....)))))...))))))((......))...........)).......((((((((...(..(....)..)..)))))))). (-28.28 = -28.24 + -0.04)

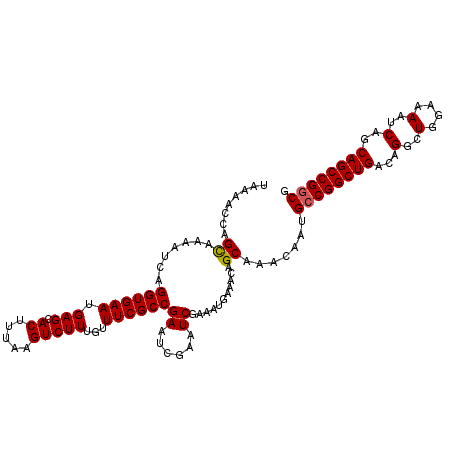
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:36 2006