
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,404,403 – 27,404,524 |
| Length | 121 |
| Max. P | 0.986542 |

| Location | 27,404,403 – 27,404,503 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -53.13 |
| Consensus MFE | -32.60 |
| Energy contribution | -32.63 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27404403 100 + 27905053 -----UCCCAGGG------CCCGGUCCCGAGGGAGGGCCCAACGGGCUGCCAAUUGGAUUGCCCAUUGGCGAACCCAUAGGCAGUGAGUUCAUUGGCGAGUUCUGCCCCGG-- -----..((.(((------(((..(((....)))))))))...((((.((((((.((....)).))))))((((.....(.(((((....))))).)..)))).)))).))-- ( -46.50) >DroGri_CAF1 7243 106 + 1 -CCGGGUCCGGGC------CCUGGUCCUGGUCCGGGACCCAGUGGACUGCCAAUCUGGUUGCCCAUGGGUGAGCCCAUGGGCAGCGAGUUCAUUGGCGAGUUCUGGCCGGGCA -((((..(.((((------....)))).)..))))(.(((.(((((((((((((((.((((((((((((....)))))))))))).))...)))))).)))))..)).)))). ( -64.60) >DroEre_CAF1 6785 100 + 1 -----UUCCCGGG------CCCGGUCCCGAAGGAGGGCCCAACGGGCUGCCAAUUGGAUUGCCCAUCGGCGAGCCCAUGGGCAGUGAGUUCAUUGGCGAGUUCUGGCCCGG-- -----...(((((------((.((((((....).)))))....(((((((((((.(((((((((((.((....)).)))))))....)))))))))).))))).)))))))-- ( -54.60) >DroYak_CAF1 6094 100 + 1 -----UUCCAGGG------CCCGGACCCGAGGGAGGGCCCAACGGGCUGCCAAUUGGAUUGCCCAUCGGCGAGCCCAUGGGCAGUGAGUUCAUUGGCGAGUUCUGGCCCGG-- -----.....(((------(((...((....)).))))))..((((((((((((.(((((((((((.((....)).)))))))....))))))))))(....).)))))).-- ( -52.30) >DroMoj_CAF1 7169 106 + 1 -CCGGGUCCAGGA------CCAGGUCCAGGUCCGGGACCUAGCGGACUGCCAAUUUGAUUGCCCAUAGGCGAACCCAUUGGCAGCGAGUUCAUAGGAGAGUUUUGCCCGGGCA -((((((((.(((------((.......))))).)).((((((...((((((((..(.(((((....))))).)..))))))))...))...))))........))))))... ( -44.50) >DroAna_CAF1 6391 113 + 1 UCCGGGUCCAGGCCCCGGACCUGGGCCGGAAGGAGGUCCUAGCGGGCUUCCAAUUGGGUUACCCAUCGGCGAACCCAUGGGUAGCGAGUUCAUAGGCGAGUUCUGGCCAGGCA ((((((.......))))))(((((.((((((((((((((....))))))))......(((((((((.((....)).)))))))))...............))))))))))).. ( -56.30) >consensus _____GUCCAGGG______CCCGGUCCCGAAGGAGGGCCCAACGGGCUGCCAAUUGGAUUGCCCAUCGGCGAACCCAUGGGCAGCGAGUUCAUUGGCGAGUUCUGGCCCGG__ .......((.((..........(((((.......)))))....(((((((((((...(((((((((.((....)).)))))))))......)))))).)))))...)).)).. (-32.60 = -32.63 + 0.03)



| Location | 27,404,403 – 27,404,503 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -48.72 |
| Consensus MFE | -26.61 |
| Energy contribution | -27.14 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27404403 100 - 27905053 --CCGGGGCAGAACUCGCCAAUGAACUCACUGCCUAUGGGUUCGCCAAUGGGCAAUCCAAUUGGCAGCCCGUUGGGCCCUCCCUCGGGACCGGG------CCCUGGGA----- --(((((((.(....)(((((((((((((.......)))))))(((....)))......)))))).))))...(((((((((....)))..)))------))))))..----- ( -47.00) >DroGri_CAF1 7243 106 - 1 UGCCCGGCCAGAACUCGCCAAUGAACUCGCUGCCCAUGGGCUCACCCAUGGGCAACCAGAUUGGCAGUCCACUGGGUCCCGGACCAGGACCAGG------GCCCGGACCCGG- ...((((...(.(((.((((((...((.(.((((((((((....)))))))))).).))))))))))).).((((((((.((.......)).))------))))))..))))- ( -52.40) >DroEre_CAF1 6785 100 - 1 --CCGGGCCAGAACUCGCCAAUGAACUCACUGCCCAUGGGCUCGCCGAUGGGCAAUCCAAUUGGCAGCCCGUUGGGCCCUCCUUCGGGACCGGG------CCCGGGAA----- --.(((((..(....)((((((........(((((((.((....)).))))))).....)))))).)))))(((((((((((....)))..)))------)))))...----- ( -53.32) >DroYak_CAF1 6094 100 - 1 --CCGGGCCAGAACUCGCCAAUGAACUCACUGCCCAUGGGCUCGCCGAUGGGCAAUCCAAUUGGCAGCCCGUUGGGCCCUCCCUCGGGUCCGGG------CCCUGGAA----- --(((((((.......((((((........(((((((.((....)).))))))).....)))))).......(((((((......)))))))))------))).))..----- ( -49.82) >DroMoj_CAF1 7169 106 - 1 UGCCCGGGCAAAACUCUCCUAUGAACUCGCUGCCAAUGGGUUCGCCUAUGGGCAAUCAAAUUGGCAGUCCGCUAGGUCCCGGACCUGGACCUGG------UCCUGGACCCGG- ...((((((.....((......))....(((((((((.((((.(((....)))))))..)))))))))..))).(((((.(((((.......))------))).))))))))- ( -44.90) >DroAna_CAF1 6391 113 - 1 UGCCUGGCCAGAACUCGCCUAUGAACUCGCUACCCAUGGGUUCGCCGAUGGGUAACCCAAUUGGAAGCCCGCUAGGACCUCCUUCCGGCCCAGGUCCGGGGCCUGGACCCGGA ..((((((..((..(((....)))..))(((((((((.((....)).)))))))..((....))..))..)))))).......(((((.((((((.....))))))..))))) ( -44.90) >consensus __CCGGGCCAGAACUCGCCAAUGAACUCACUGCCCAUGGGCUCGCCGAUGGGCAAUCCAAUUGGCAGCCCGCUGGGCCCUCCAUCGGGACCGGG______CCCUGGAA_____ ..((.(((........((((((........(((((((.((....)).))))))).....)))))).........((.((.......)).)).........))).))....... (-26.61 = -27.14 + 0.53)

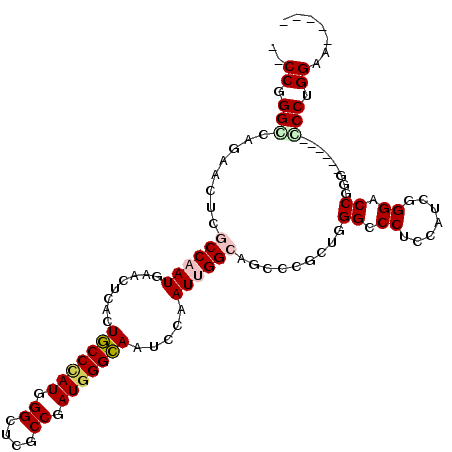

| Location | 27,404,428 – 27,404,524 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.00 |
| Mean single sequence MFE | -45.52 |
| Consensus MFE | -16.64 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27404428 96 + 27905053 GCCCAACGGGCUGCCAAUUGGAUUGCCCAUUGGCGAACCCAUAGGCAGUGAGUUCAUUGGCGAGUUCUGCCCCGG------ACCGC---------CCAUCGGUGAACCCUG .......((((((((.((.((.(((((....))))).)).)).))))))..(((((((((((.((((......))------)))))---------)....))))))))).. ( -37.50) >DroVir_CAF1 7502 111 + 1 ACCCAGUGGACUGCCAAUUUGAUUGCCCAUGGGCGAGCCCAUUGGCAGCGAGUUCAUUGGCGAAUUCUGUCCGGGCAUCGGUCCACCAGCACCGCCCAUGGGCGAUCCCUG ...((((((....)))....(((((((((((((((.((((...((((..((((((......)))))))))).))))...((....)).....))))))))))))))).))) ( -51.30) >DroPse_CAF1 7126 105 + 1 UCCAAGGGGACUACCAAUGGGAUUACCCAUCGGCGAGCCCAUCGGCAACGAAUUCAUGGGCGAGUUCUGGCCAGG------ACCGCCAGCUCCUCCCAUGGGAGAGCCCUG .....((((.((.((.(((((....))))).)).((((...(((....))).......((((.(((((....)))------)))))).))))((((....)))))))))). ( -44.50) >DroGri_CAF1 7272 111 + 1 ACCCAGUGGACUGCCAAUCUGGUUGCCCAUGGGUGAGCCCAUGGGCAGCGAGUUCAUUGGCGAGUUCUGGCCGGGCAUGGGUCCACCAGCGCCGCCCAUGGGCGAGCCUUG .......(((((((((((((.((((((((((((....)))))))))))).))...)))))).))))).((((.(((.(((.....)))..)))(((....)))).)))... ( -60.50) >DroAna_CAF1 6427 99 + 1 UCCUAGCGGGCUUCCAAUUGGGUUACCCAUCGGCGAACCCAUGGGUAGCGAGUUCAUAGGCGAGUUCUGGCCAGGCAC---ACCUC---------CCAUUGGAGAUCCUUG .......((..(((((((.(((((((((((.((....)).))))))))).........(((.(....).)))(((...---.))))---------).)))))))..))... ( -36.40) >DroPer_CAF1 6571 105 + 1 UCCAAGGGGACUACCAAUGGGAUUACCCAUCGGCGAGCCCAUCGGCAACGAAUUCAUGGGCGAGUUUUGGCCAGG------ACCGCCAGCUCCUCCCAUGGGAGAGCCCUG ......(((.((.((.(((((....))))).))..))))).(((....)))......((((((((..((((....------...))))))))((((....)))).)))).. ( -42.90) >consensus UCCCAGCGGACUGCCAAUUGGAUUACCCAUCGGCGAGCCCAUCGGCAGCGAGUUCAUUGGCGAGUUCUGGCCAGG______ACCGCCAGC_CC_CCCAUGGGAGAGCCCUG .......((..((((.......(((((.((.((....)).)).))))).((((((......)))))).................................))))..))... (-16.64 = -16.20 + -0.44)



| Location | 27,404,428 – 27,404,524 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.00 |
| Mean single sequence MFE | -46.38 |
| Consensus MFE | -20.27 |
| Energy contribution | -21.97 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27404428 96 - 27905053 CAGGGUUCACCGAUGG---------GCGGU------CCGGGGCAGAACUCGCCAAUGAACUCACUGCCUAUGGGUUCGCCAAUGGGCAAUCCAAUUGGCAGCCCGUUGGGC .........(((((((---------((...------..(((......)))(((((((((((((.......)))))))(((....)))......)))))).))))))))).. ( -40.80) >DroVir_CAF1 7502 111 - 1 CAGGGAUCGCCCAUGGGCGGUGCUGGUGGACCGAUGCCCGGACAGAAUUCGCCAAUGAACUCGCUGCCAAUGGGCUCGCCCAUGGGCAAUCAAAUUGGCAGUCCACUGGGU (((((((.(((((((((((((.(....).)))((.((((((.(((..((((....))))....)))))...))))))))))))))))..((.....))..)))).)))... ( -48.20) >DroPse_CAF1 7126 105 - 1 CAGGGCUCUCCCAUGGGAGGAGCUGGCGGU------CCUGGCCAGAACUCGCCCAUGAAUUCGUUGCCGAUGGGCUCGCCGAUGGGUAAUCCCAUUGGUAGUCCCCUUGGA ..((((.((((....))))((((((((...------....)))))..)))))))............((((.(((((.(((((((((....))))))))))).))).)))). ( -51.20) >DroGri_CAF1 7272 111 - 1 CAAGGCUCGCCCAUGGGCGGCGCUGGUGGACCCAUGCCCGGCCAGAACUCGCCAAUGAACUCGCUGCCCAUGGGCUCACCCAUGGGCAACCAGAUUGGCAGUCCACUGGGU ....((.((((....)))))).(..((((((...((((.(((........)))......((.(.((((((((((....)))))))))).).))...))))))))))..).. ( -55.20) >DroAna_CAF1 6427 99 - 1 CAAGGAUCUCCAAUGG---------GAGGU---GUGCCUGGCCAGAACUCGCCUAUGAACUCGCUACCCAUGGGUUCGCCGAUGGGUAACCCAAUUGGAAGCCCGCUAGGA ...((...((((((((---------(((((---(.(.((....))..).)))))..........(((((((.((....)).))))))).))).))))))...))....... ( -34.40) >DroPer_CAF1 6571 105 - 1 CAGGGCUCUCCCAUGGGAGGAGCUGGCGGU------CCUGGCCAAAACUCGCCCAUGAAUUCGUUGCCGAUGGGCUCGCCGAUGGGUAAUCCCAUUGGUAGUCCCCUUGGA ..((((.((((....))))(((.((((...------....))))...)))))))............((((.(((((.(((((((((....))))))))))).))).)))). ( -48.50) >consensus CAGGGCUCUCCCAUGGG_GG_GCUGGCGGU______CCUGGCCAGAACUCGCCAAUGAACUCGCUGCCCAUGGGCUCGCCGAUGGGCAAUCCAAUUGGCAGUCCACUGGGA ...(((.((((....))))........((.......))............))).............((((((((((.((((((.((....)).)))))))))))).)))). (-20.27 = -21.97 + 1.70)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:51:48 2006