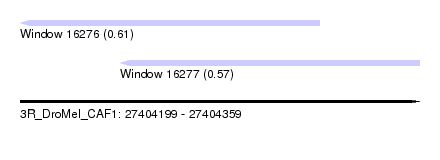
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,404,199 – 27,404,359 |
| Length | 160 |
| Max. P | 0.611103 |

| Location | 27,404,199 – 27,404,319 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.13 |
| Mean single sequence MFE | -54.55 |
| Consensus MFE | -35.21 |
| Energy contribution | -35.77 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27404199 120 - 27905053 CUCUGCACAGUCCGCUCGGAAACGGACCAACGGGUCAUGGCAGUCACAUGCCUGGAGGACCAAUCCCAGGACCAGGUCCUGGGCCUGGCGGCCUAGUAGGUCCCGGUGGCAUCUCCCCCG .........(((((........)))))...((((....((((......)))).(((((.(((..((((((((...)))))))).((((.((((.....)))))))))))..))))))))) ( -50.90) >DroVir_CAF1 7273 117 - 1 CCCUGCACAGUCCACUGGGUAAUGGGCCAACAAAUCA---CAGUCACAUGUCCGGUGGACCAGGUCCAGGCCCGGGUCCUGGGCCGGGCGGUUUAGUUGGACCCGGUGGCAUGUCGCCAG .((((.((.(((((((((..(.(((((..........---..))).)))..)))))))))...)).)))).((((((((((((((....))))))...))))))))((((.....)))). ( -52.90) >DroPse_CAF1 6885 120 - 1 CGUUGCACAGUCCGCUGGGCAAUGGACCGACGAAUCAUGGCAGUCAUAUGCCAGGUGGACCCGGUCCAGGUCCUGGGCCUGGGCCCGGUGGCCUCGUCGGACCAGGUGGCAUGUCUCCCG .(((((.(((....))).)))))((.(((((((....(((((......)))))(((.(.((.(((((((((.....))))))))).))).)))))))))).)).((.((....)).)).. ( -58.90) >DroWil_CAF1 6233 120 - 1 CUUUGCACAGUCCUCUUGGCAAUGGUCCUGCGAGUCAUGGCAAUCAUAUGCCUGGUGGGCCAGGUCCGGGACCAGGUCCUGGGCCAGGCAAUCUAGUCGGGCCUGGAGGCAUUUCUCCCG .........(.(((((.(((..((((((.((.((.((((.......)))).)).))))))))((((((((((...)))))))))).(((......)))..))).)))))).......... ( -49.20) >DroMoj_CAF1 6951 117 - 1 CUCUGCACAGUCCGCUGGGCAAUGGACCAACGAAUCA---CAGUCAUAUGUCCGGCGGACCAGGUCCGGGGCCAGGUCCUGGGCCGGGUGGCUUAGUCGGACCUGGUGGCAUGUCGCCGG .((((.((.((((((((((((...(((..........---..)))...))))))))))))...)).))))(((((((((((((((....))))))...)))))))))(((.....))).. ( -57.00) >DroAna_CAF1 6180 120 - 1 CACUGCAUAGUCCGUUGGGCAACGGUCCCACGGGUCACGGGAGUCACAUGCCCGGCGGACCUGGUCCCGGGCCUGGACCUGGGCCUGGUGGCGUCGUGGGACCUGGUGGCAUAUCCCCCG ((((.....((((...))))...(((((((((((((.((((.........))))...))))..(((((((((((......)))))))).)))..))))))))).))))............ ( -58.40) >consensus CUCUGCACAGUCCGCUGGGCAAUGGACCAACGAAUCAUGGCAGUCACAUGCCCGGUGGACCAGGUCCAGGACCAGGUCCUGGGCCGGGCGGCCUAGUCGGACCUGGUGGCAUGUCCCCCG .........(.((((..(((......(((((.......((((......)))).(((.(.((.((((((((((...)))))))))).))).)))..))))))))..))))).......... (-35.21 = -35.77 + 0.56)



| Location | 27,404,239 – 27,404,359 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -45.27 |
| Consensus MFE | -30.93 |
| Energy contribution | -31.63 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27404239 120 - 27905053 CCGCAGCCACCGAUGAACAACGGGCAGAUGGGUCCUCCUCCUCUGCACAGUCCGCUCGGAAACGGACCAACGGGUCAUGGCAGUCACAUGCCUGGAGGACCAAUCCCAGGACCAGGUCCU (((.(((..(((........)))(((((.(((......)))))))).......))))))....(((((..((((.((((.......))))))))..((.((.......)).)).))))). ( -43.40) >DroVir_CAF1 7313 117 - 1 CAACAGACGCACAUGAACAAUGGACAAAUGGGUCCGCCGCCCCUGCACAGUCCACUGGGUAAUGGGCCAACAAAUCA---CAGUCACAUGUCCGGUGGACCAGGUCCAGGCCCGGGUCCU .....(((.............((((......)))).(((..((((.((.(((((((((..(.(((((..........---..))).)))..)))))))))...)).))))..)))))).. ( -39.90) >DroPse_CAF1 6925 120 - 1 CCGCAGCCACCCAUGAACAAUGGACAGAUGGGUCCGCCCCCGUUGCACAGUCCGCUGGGCAAUGGACCGACGAAUCAUGGCAGUCAUAUGCCAGGUGGACCCGGUCCAGGUCCUGGGCCU .........((((.((....(((((....(((((((((.(((((((.(((....))).)))))))............(((((......)))))))))))))).)))))..)).))))... ( -56.10) >DroWil_CAF1 6273 120 - 1 CCGCAGCCACCCAUGAAUAAUGGACAAAUGGGUCCUCCUCCUUUGCACAGUCCUCUUGGCAAUGGUCCUGCGAGUCAUGGCAAUCAUAUGCCUGGUGGGCCAGGUCCGGGACCAGGUCCU .(((((..((((((.............))))))......((.((((.(((.....))))))).))..)))))..(((.((((......))))))).(((((.((((...)))).))))). ( -39.02) >DroMoj_CAF1 6991 117 - 1 CAACAGACGCACAUGAAUAAUGGACAAAUGGGUCCGCCGCCUCUGCACAGUCCGCUGGGCAAUGGACCAACGAAUCA---CAGUCAUAUGUCCGGCGGACCAGGUCCGGGGCCAGGUCCU .....................((((...(((.((((((((....))...((((((((((((...(((..........---..)))...))))))))))))..))..)))).))).)))). ( -44.80) >DroAna_CAF1 6220 120 - 1 CCGCAGCCACCGAUGAACAAUGGACAGAUGGGUCCGCCGCCACUGCAUAGUCCGUUGGGCAACGGUCCCACGGGUCACGGGAGUCACAUGCCCGGCGGACCUGGUCCCGGGCCUGGACCU (((..(((((((.((..((((((((.....((....))((....))...))))))))..)).))))......)))..)))..(((.((.((((((..(((...))))))))).))))).. ( -48.40) >consensus CCGCAGCCACCCAUGAACAAUGGACAAAUGGGUCCGCCGCCUCUGCACAGUCCGCUGGGCAAUGGACCAACGAAUCAUGGCAGUCACAUGCCCGGUGGACCAGGUCCAGGACCAGGUCCU .....................((((.....((((((((.((..(((.(((....))).)))..)).............((((......)))).))))))))..))))(((((...))))) (-30.93 = -31.63 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:51:45 2006