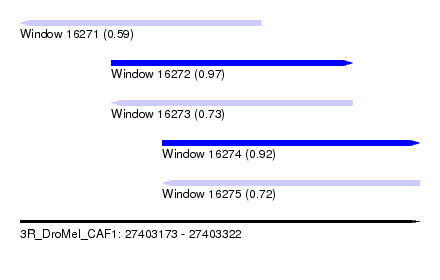
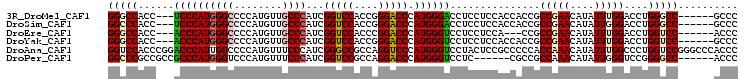
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,403,173 – 27,403,322 |
| Length | 149 |
| Max. P | 0.972291 |

| Location | 27,403,173 – 27,403,263 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -38.23 |
| Consensus MFE | -22.92 |
| Energy contribution | -22.95 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27403173 90 - 27905053 GACCGAUGGGCAACAUGGGGCCCAUGGGA---GGUGGCCCGAUGGGCGGC------CCUAUGGGCGUAGGUCCCAAGCCGAUGACAAUGGGCGGCGGGA ..((.(((.....))).))(((((((((.---.((.((((...)))).))------))))))))).....((((..(((.((....)).)))...)))) ( -44.50) >DroPse_CAF1 5727 93 - 1 GACCGAUGGGAAACAUGGGACCCAUGGGCGGCGGCGGGCCAAUGGGUGGA------CCGAUGGGAGUGGGACCCAAGCCGAUGACAAUGAGCGGUGGCA ..((.(((.....))).))...(((.((((((.....)))..(((((...------((.((....)).))))))).))).))).......((....)). ( -30.20) >DroEre_CAF1 5561 90 - 1 GACCGAUGGGCAACAUGGGGCCCAUGGGU---GGUGGCCCGAUGGGCGGC------CCCAUGGGUGUAGGUCCCAAGCCGAUGACAAUGGGCGGCGGAA ..(((.((((..((...(.(((((((((.---.((.((((...)))).))------))))))))).)..)))))).(((.((....)).)))..))).. ( -44.30) >DroWil_CAF1 5210 93 - 1 GUCCUAUGGGGAAUAUGGGCCCAAUGAGC------GGUCCAAUGGGUGGCGUAGGGCCAAUGGGCGUAGGGCCCAAGCCGAUGACAAUGAGUGGCGGUA .(((.....)))...(((((((.(((...------(((((.(((.....))).)))))......))).))))))).((((.(.((.....)).))))). ( -34.60) >DroAna_CAF1 5154 93 - 1 GCCCGAUGGGAAACAUGGGGCCAAUGGGUCCGGGUGGACCUAUGGGAGGG------CCCAUGGGCGUGGGGCCCAAGCCGAUGACAAUGGGUGGCGGCA ((((.(((.....))).))))...(((((((.(...(.((((((((....------))))))))).).))))))).((((.(.((.....)).))))). ( -45.60) >DroPer_CAF1 5184 93 - 1 GACCGAUGGGAAACAUGGGACCCAUGGGCGGCGGCGGGCCAAUGGGUGGA------CCGAUGGGAGUGGGACCCAAGCCGAUGACAAUGAGCGGUGGCA ..((.(((.....))).))...(((.((((((.....)))..(((((...------((.((....)).))))))).))).))).......((....)). ( -30.20) >consensus GACCGAUGGGAAACAUGGGGCCCAUGGGC___GGUGGCCCAAUGGGUGGC______CCCAUGGGCGUAGGACCCAAGCCGAUGACAAUGAGCGGCGGCA ..((.(((.....))).))((((((.((..........)).)))))).........((..((((.......)))).((((...........)))))).. (-22.92 = -22.95 + 0.03)


| Location | 27,403,207 – 27,403,297 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 88.79 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -29.16 |
| Energy contribution | -29.77 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.51 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27403207 90 + 27905053 GCCCAUAGGGCCGCCCAUCGGGCCACC---UCCCAUGGGCCCCAUGUUGCCCAUCGGUCCACCGGGACCCAUGGGACCUCCUCCACCACCGCC ((.(((.(((..((((((.(((.....---.)))))))))))))))...(((((.(((((....))))).)))))...............)). ( -37.10) >DroSec_CAF1 5332 90 + 1 GCCCAUAGGGCCACCCAUCGGCCCACC---UCCCAUGGGCCCCAUGUUGCCCAUCGGUCCACCGGGUCCCAUGGGACCUCCUCCACCACCGCC ((.....(((((.......)))))...---((((((((((((..((..(((....))).))..))).)))))))))..............)). ( -36.90) >DroSim_CAF1 4949 90 + 1 GCCCAUAGGGCCGCCCAUCGGCCCACC---UCCCAUGGGCCCCAUGUUGCCCAUCGGUCCACCGGGACCCAUGGGACCUCCUCCACCACCGCC ((.....((((((.....))))))...---((((((((((((..((..(((....))).))..))).)))))))))..............)). ( -38.60) >DroEre_CAF1 5595 87 + 1 ACCCAUGGGGCCGCCCAUCGGGCCACC---ACCCAUGGGCCCCAUGUUGCCCAUCGGUCCACCCGGACCCAUGGGUCCUCCUCCA---CCGCC ...(((((((((((((...))))...(---(....)))))))))))..((((((.(((((....))))).)))))).........---..... ( -41.60) >DroYak_CAF1 4898 90 + 1 ACCCAUUGGGCCGCCCAUCGGGCCACC---ACCCAUGGGCCCCAUGUUGCCCAUCGGUCCACCGGGACCCAUGGGUCCUCCUCCACCACCGCC ......((((..((((((.(((.....---.)))))))))))))....((((((.(((((....))))).))))))................. ( -38.00) >DroAna_CAF1 5188 93 + 1 GCCCAUGGGCCCUCCCAUAGGUCCACCCGGACCCAUUGGCCCCAUGUUUCCCAUCGGGCCGCCAGGUCCCAUGGGUCCUACUCCGCCCCCACC (((((((((.(((..(...(((((....)))))....(((((.(((.....))).))))))..))).)))))))))................. ( -41.70) >consensus GCCCAUAGGGCCGCCCAUCGGGCCACC___ACCCAUGGGCCCCAUGUUGCCCAUCGGUCCACCGGGACCCAUGGGACCUCCUCCACCACCGCC ...(((.(((..((((((.((..........)).))))))))))))...(((((.(((((....))))).))))).................. (-29.16 = -29.77 + 0.61)



| Location | 27,403,207 – 27,403,297 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 88.79 |
| Mean single sequence MFE | -45.87 |
| Consensus MFE | -35.04 |
| Energy contribution | -35.35 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27403207 90 - 27905053 GGCGGUGGUGGAGGAGGUCCCAUGGGUCCCGGUGGACCGAUGGGCAACAUGGGGCCCAUGGGA---GGUGGCCCGAUGGGCGGCCCUAUGGGC (.(.((((.........((((((((((((((((.(.((....))).)).))))))))))))))---(((.((((...)))).))))))).).) ( -47.70) >DroSec_CAF1 5332 90 - 1 GGCGGUGGUGGAGGAGGUCCCAUGGGACCCGGUGGACCGAUGGGCAACAUGGGGCCCAUGGGA---GGUGGGCCGAUGGGUGGCCCUAUGGGC (.(.(((..........(((((((((.(((.(((..((....))...))).))))))))))))---...((((((.....))))))))).).) ( -42.10) >DroSim_CAF1 4949 90 - 1 GGCGGUGGUGGAGGAGGUCCCAUGGGUCCCGGUGGACCGAUGGGCAACAUGGGGCCCAUGGGA---GGUGGGCCGAUGGGCGGCCCUAUGGGC (.(.(((..........((((((((((((((((.(.((....))).)).))))))))))))))---...((((((.....))))))))).).) ( -45.60) >DroEre_CAF1 5595 87 - 1 GGCGG---UGGAGGAGGACCCAUGGGUCCGGGUGGACCGAUGGGCAACAUGGGGCCCAUGGGU---GGUGGCCCGAUGGGCGGCCCCAUGGGU ..(.(---((......(.(((((.(((((....))))).))))))..))).).(((((((((.---.((.((((...)))).))))))))))) ( -46.90) >DroYak_CAF1 4898 90 - 1 GGCGGUGGUGGAGGAGGACCCAUGGGUCCCGGUGGACCGAUGGGCAACAUGGGGCCCAUGGGU---GGUGGCCCGAUGGGCGGCCCAAUGGGU ..((.((..........((((((((((((((((.(.((....))).)).))))))))))))))---(((.((((...)))).))))).))... ( -43.80) >DroAna_CAF1 5188 93 - 1 GGUGGGGGCGGAGUAGGACCCAUGGGACCUGGCGGCCCGAUGGGAAACAUGGGGCCAAUGGGUCCGGGUGGACCUAUGGGAGGGCCCAUGGGC .(((((..(......)..))))).(((((((..(((((.(((.....))).)))))..))))))).......((((((((....)))))))). ( -49.10) >consensus GGCGGUGGUGGAGGAGGACCCAUGGGUCCCGGUGGACCGAUGGGCAACAUGGGGCCCAUGGGA___GGUGGCCCGAUGGGCGGCCCUAUGGGC .((...............(((((.(((((....))))).)))))...((((((((((((.((..........)).)))...))))))))).)) (-35.04 = -35.35 + 0.31)

| Location | 27,403,226 – 27,403,322 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.22 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -29.83 |
| Energy contribution | -29.88 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27403226 96 + 27905053 GGGCCACC---UCCCAUGGGCCCCAUGUUGCCCAUCGGUCCACCGGGACCCAUGGGACCUCCUCCACCACCGCCGAACAUAUUUGGACCUGGGCC------GCCC ((((....---((((((((((((..((..(((....))).))..))).))))))))).........(((...(((((....)))))...)))...------)))) ( -38.90) >DroSim_CAF1 4968 96 + 1 GGCCCACC---UCCCAUGGGCCCCAUGUUGCCCAUCGGUCCACCGGGACCCAUGGGACCUCCUCCACCACCGCCGAACAUAUUGGGACCUGGGCC------GCCC ((((((..---((((((((((........)))))(((((((....)))))..((((......))))........))......)))))..))))))------.... ( -40.20) >DroEre_CAF1 5614 93 + 1 GGGCCACC---ACCCAUGGGCCCCAUGUUGCCCAUCGGUCCACCCGGACCCAUGGGUCCUCCUCCA---CCGCCGAACAUAUUUGGACCUGGUCC------ACCC ((((((..---....((((((........)))))).((((((...(((((....)))))...((..---.....)).......))))))))))))------.... ( -36.40) >DroYak_CAF1 4917 96 + 1 GGGCCACC---ACCCAUGGGCCCCAUGUUGCCCAUCGGUCCACCGGGACCCAUGGGUCCUCCUCCACCACCGCCGAACAUAUUUGGACCUGGUCC------GCCC ((((....---((((((((((((..((..(((....))).))..))).)))))))))........((((...(((((....)))))...))))..------)))) ( -38.80) >DroAna_CAF1 5207 105 + 1 GGUCCACCCGGACCCAUUGGCCCCAUGUUUCCCAUCGGGCCGCCAGGUCCCAUGGGUCCUACUCCGCCCCCACCAAACAUAUUUGGCCCUGGUCCGGGCCCACCC (((...((((((((....((((..(((((((((((.(((((....))))).)))))..................))))))....))))..))))))))...))). ( -44.16) >DroPer_CAF1 5237 93 + 1 GGCCCGCCGCCGCCCAUGGGUCCCAUGUUUCCCAUCGGUCCGCCAGGACCCAUGGGUCCUC------CGCCGCCAAACAUAUUGGGUCCGGGGCC------ACCC (((((((.(..((((((((((((.(((.....))).((....)).))))))))))))...)------.))..((((.....)))).....)))))------.... ( -41.40) >consensus GGGCCACC___ACCCAUGGGCCCCAUGUUGCCCAUCGGUCCACCGGGACCCAUGGGUCCUCCUCCACCACCGCCGAACAUAUUUGGACCUGGGCC______ACCC (((((......((((((((((........))))...(((((....))))).))))))...............(((((....)))))....))))).......... (-29.83 = -29.88 + 0.06)
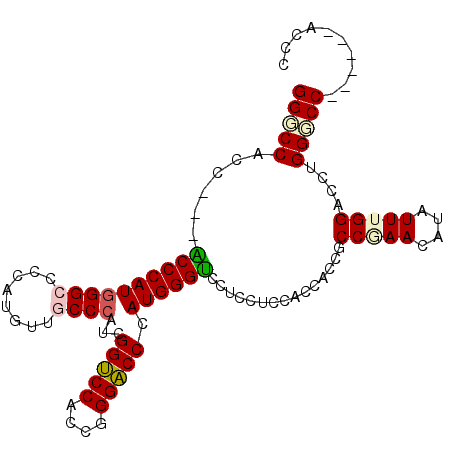
| Location | 27,403,226 – 27,403,322 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.22 |
| Mean single sequence MFE | -48.52 |
| Consensus MFE | -30.50 |
| Energy contribution | -31.42 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27403226 96 - 27905053 GGGC------GGCCCAGGUCCAAAUAUGUUCGGCGGUGGUGGAGGAGGUCCCAUGGGUCCCGGUGGACCGAUGGGCAACAUGGGGCCCAUGGGA---GGUGGCCC ((((------.(((....((((....((.....))....)))).....((((((((((((((((.(.((....))).)).))))))))))))))---))).)))) ( -49.70) >DroSim_CAF1 4968 96 - 1 GGGC------GGCCCAGGUCCCAAUAUGUUCGGCGGUGGUGGAGGAGGUCCCAUGGGUCCCGGUGGACCGAUGGGCAACAUGGGGCCCAUGGGA---GGUGGGCC ....------((((((..(((((...(.(((.((....)).))).)(((((((((.(((((((....)))..))))..)))))))))..)))))---..)))))) ( -49.90) >DroEre_CAF1 5614 93 - 1 GGGU------GGACCAGGUCCAAAUAUGUUCGGCGG---UGGAGGAGGACCCAUGGGUCCGGGUGGACCGAUGGGCAACAUGGGGCCCAUGGGU---GGUGGCCC ((((------.(.((...((((....((.....)).---)))).....((((((((((((((.....))....(....)...))))))))))))---))).)))) ( -41.20) >DroYak_CAF1 4917 96 - 1 GGGC------GGACCAGGUCCAAAUAUGUUCGGCGGUGGUGGAGGAGGACCCAUGGGUCCCGGUGGACCGAUGGGCAACAUGGGGCCCAUGGGU---GGUGGCCC ((((------.(.((...((((....((.....))....)))).....((((((((((((((((.(.((....))).)).))))))))))))))---))).)))) ( -46.50) >DroAna_CAF1 5207 105 - 1 GGGUGGGCCCGGACCAGGGCCAAAUAUGUUUGGUGGGGGCGGAGUAGGACCCAUGGGACCUGGCGGCCCGAUGGGAAACAUGGGGCCAAUGGGUCCGGGUGGACC .(((..(((((((((...((((.....((((.(((((..(......)..))))).)))).))))(((((.(((.....))).)))))....)))))))))..))) ( -58.10) >DroPer_CAF1 5237 93 - 1 GGGU------GGCCCCGGACCCAAUAUGUUUGGCGGCG------GAGGACCCAUGGGUCCUGGCGGACCGAUGGGAAACAUGGGACCCAUGGGCGGCGGCGGGCC ....------((((((((((.......)))))((.((.------(....((((((((((((((....))....(....)..))))))))))))).)).))))))) ( -45.70) >consensus GGGC______GGACCAGGUCCAAAUAUGUUCGGCGGUGGUGGAGGAGGACCCAUGGGUCCCGGUGGACCGAUGGGCAACAUGGGGCCCAUGGGU___GGUGGCCC ..........((.(((...((.......(((.((....)).)))....(((((((((((((((....))....(....)..)))))))))))))...))))).)) (-30.50 = -31.42 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:51:43 2006