
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,401,045 – 27,401,193 |
| Length | 148 |
| Max. P | 0.850763 |

| Location | 27,401,045 – 27,401,165 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -40.89 |
| Consensus MFE | -22.58 |
| Energy contribution | -23.37 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27401045 120 + 27905053 AAUCUGACGGUGCGCAGCACGGACCAGGACGUGGCGGAGCUGAUAACGGCGGCCAUUGGAUAAGACAGCGAGCCAUGCAGGAACCUCCUUUGGUUCCUGCGUGCUGUGAACGUAAUCUGA ......(((...((((((((..........(((((((.((((....))))..)).(((.......)))...)))))(((((((((......)))))))))))))))))..)))....... ( -51.10) >DroVir_CAF1 3122 120 + 1 AAUCUAACCGUGCGUAGCACAGAUCAGGAUGUGGCCGAGCUCAUAACGGCAGCCAUCGGAUAAACCUACGCGCCAUGCAGAGACUUAAUAGGAUCUCUGCGUGCUCUGCAAAUACUAUGA .........((((((((.....(((..((((..((((.........))))...)))).)))....))))))))((((((((((((.....)).))))))))))................. ( -40.40) >DroGri_CAF1 3378 120 + 1 AAUCUGACAGUGCGUAGCACGGAUCAGGAUGUGGCCGAGCUCAUAACGGCAGCCAUCGGAUAAGAUUGCGAGCCAUGCAGAGAUCUCGUUGGGUUUCUGCGUGCCAUGAAUAUGAUAUGA ..(((((..((((...))))...)))))..(((((............(((.((.(((......))).))..)))((((((((((((....)))))))))))))))))............. ( -40.50) >DroWil_CAF1 2823 120 + 1 AAUCUAACUGUGCGAAGCACAGAUCAAGAUGUAGCAGAGGUUAUAACGGCGGCCAUUGGAUAAGACAGCGAGCCAUGCAGAAAUCUUGUUGGUUUUCUGCGUGCAAUUAACAAAAUAUAA .......((((((...)))))).....(.((((((....)))))).).((((((.(((.......))).).)))(((((((((.((....)).)))))))))))................ ( -31.10) >DroMoj_CAF1 3257 120 + 1 AAUUUAACCGUGCGAAGCACAGAUCAGGAUGUGGCCGAGCUAAUAACGGCAGCGAUCGGAUAAAAUUGCGCACCAUGCAGAGACCUAAUCGUACAGCUGCGUACUCUGCAUAUAUUAUGA .........(((((........(((..(((((.((((.........)))).)).))).))).......))))).((((((((.......((((....))))..))))))))......... ( -31.06) >DroAna_CAF1 3210 120 + 1 AACCUGACCGUGCGCAGCACAGAUCAGGAUGUGGCGGAGCUAAUAACGGCGGCCAUCGGAUAAGACAGCGCGCCAUGCAGGAAUCUGAUAUGGUUCCUGCGUGCUGCCAACAUAAUCUGA ..((((((.((((...)))).).)))))(((((((((.((.......((((((..((......))..)).))))..(((((((((......))))))))))).))))).))))....... ( -51.20) >consensus AAUCUAACCGUGCGAAGCACAGAUCAGGAUGUGGCCGAGCUAAUAACGGCAGCCAUCGGAUAAGACAGCGAGCCAUGCAGAAACCUAAUUGGGUUUCUGCGUGCUAUGAACAUAAUAUGA .((((((..((((...))))...........((((...(((......))).))))))))))............((((((((((.(......).))))))))))................. (-22.58 = -23.37 + 0.78)



| Location | 27,401,045 – 27,401,165 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -36.94 |
| Consensus MFE | -24.05 |
| Energy contribution | -23.80 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27401045 120 - 27905053 UCAGAUUACGUUCACAGCACGCAGGAACCAAAGGAGGUUCCUGCAUGGCUCGCUGUCUUAUCCAAUGGCCGCCGUUAUCAGCUCCGCCACGUCCUGGUCCGUGCUGCGCACCGUCAGAUU ...(((...((.(.(((((((((((((((......)))))))))(((((..(((((........))))).)))))..........((((.....))))..)))))).).)).)))..... ( -41.60) >DroVir_CAF1 3122 120 - 1 UCAUAGUAUUUGCAGAGCACGCAGAGAUCCUAUUAAGUCUCUGCAUGGCGCGUAGGUUUAUCCGAUGGCUGCCGUUAUGAGCUCGGCCACAUCCUGAUCUGUGCUACGCACGGUUAGAUU ...........((.((((..((((((((........))))))))((((((.((.((.....))....)))))))).....)))).))......((((.((((((...))))))))))... ( -40.20) >DroGri_CAF1 3378 120 - 1 UCAUAUCAUAUUCAUGGCACGCAGAAACCCAACGAGAUCUCUGCAUGGCUCGCAAUCUUAUCCGAUGGCUGCCGUUAUGAGCUCGGCCACAUCCUGAUCCGUGCUACGCACUGUCAGAUU ..........((((((((..(((((..............)))))..(((..((.(((......))).)).)))))))))))............(((((..((((...)))).)))))... ( -30.44) >DroWil_CAF1 2823 120 - 1 UUAUAUUUUGUUAAUUGCACGCAGAAAACCAACAAGAUUUCUGCAUGGCUCGCUGUCUUAUCCAAUGGCCGCCGUUAUAACCUCUGCUACAUCUUGAUCUGUGCUUCGCACAGUUAGAUU ................(((.(((((((.(......).)))))))(((((..(((((........))))).))))).........)))...((((....((((((...))))))..)))). ( -28.60) >DroMoj_CAF1 3257 120 - 1 UCAUAAUAUAUGCAGAGUACGCAGCUGUACGAUUAGGUCUCUGCAUGGUGCGCAAUUUUAUCCGAUCGCUGCCGUUAUUAGCUCGGCCACAUCCUGAUCUGUGCUUCGCACGGUUAAAUU ...((((.(((((((((((((....)))))(((...)))))))))))(((((.........((((..((((.......)))))))).((((........))))...))))).)))).... ( -29.10) >DroAna_CAF1 3210 120 - 1 UCAGAUUAUGUUGGCAGCACGCAGGAACCAUAUCAGAUUCCUGCAUGGCGCGCUGUCUUAUCCGAUGGCCGCCGUUAUUAGCUCCGCCACAUCCUGAUCUGUGCUGCGCACGGUCAGGUU .......((((.((((((..(((((((.(......).)))))))((((((.((((((......)))))))))))).....)))..)))))))(((((.((((((...))))))))))).. ( -51.70) >consensus UCAUAUUAUGUUCACAGCACGCAGAAACCCAAUGAGAUCUCUGCAUGGCGCGCAGUCUUAUCCGAUGGCCGCCGUUAUGAGCUCGGCCACAUCCUGAUCUGUGCUACGCACGGUCAGAUU ................((..(((((((..........)))))))(((((..((..((......))..)).)))))..........))......((((.((((((...))))))))))... (-24.05 = -23.80 + -0.25)



| Location | 27,401,085 – 27,401,193 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.12 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -24.65 |
| Energy contribution | -24.48 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
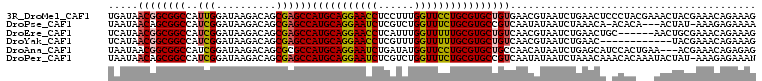
>3R_DroMel_CAF1 27401085 108 + 27905053 UGAUAACGGCGGCCAUUGGAUAAGACAGCGAGCCAUGCAGGAACCUCCUUUGGUUCCUGCGUGCUGUGAACGUAAUCUGAACUCCCUACGAAACUACGAAACAGAAAG .....(((((((((.(((.......))).).)))(((((((((((......))))))))))))))))...((((............)))).................. ( -33.90) >DroPse_CAF1 3227 103 + 1 UAAUAACAGCGGCCAUCGGAUAAGACAGCGAGCCAUGCAGGAAUCUCGUCUGGUUUCUGCGUGCCGUCAAUAUAAUCUAAACA-ACACA---ACUAU-AAAGAGAAAA ........(((((....((....(....)...))(((((((((((......))))))))))))))))................-.....---.....-.......... ( -22.30) >DroEre_CAF1 3278 102 + 1 UCAUAACGGCGGCCAUCGGAUAAGACAGCGAGCCAUGCAGGAACCUCAUUUGGUUUUUGCGUGCUGUCAACGUAAUCUGAACUGC------AACUGCGAAACAGAAAG ........((((...((((((..((((((.....(((((((((((......)))))))))))))))))......)))))).))))------..(((.....))).... ( -32.30) >DroYak_CAF1 2840 96 + 1 UCAUAACGGCGGCCAUCGGAUAAGACAGCGAGCCAUGCAGGAACCUCGUUUGGUUUUUGCGUGCUGUCAACGUAAUCUGAAC------------UACGAAACAGAAAG .......(((.((..((......))..))..)))(((((((((((......))))))))))).((((...((((........------------))))..)))).... ( -28.50) >DroAna_CAF1 3250 105 + 1 UAAUAACGGCGGCCAUCGGAUAAGACAGCGCGCCAUGCAGGAAUCUGAUAUGGUUCCUGCGUGCUGCCAACAUAAUCUGAGCAUCCACUGAA---ACGAAACAGAGAG ......(((.((...((((((......(.(((.((((((((((((......)))))))))))).))))......))))))....)).)))..---............. ( -34.30) >DroPer_CAF1 2566 107 + 1 UAAUAACAGCGGCCAUCGGAUAAGACAGCGAGCCAUGCAGGAAUCUCGUCUGGUUUCUGCGUGCCGUCAAUAUAAUCUAAACAAACACAAAUACUAU-AAAGAGAAAN ........(((((....((....(....)...))(((((((((((......))))))))))))))))..............................-.......... ( -22.30) >consensus UAAUAACGGCGGCCAUCGGAUAAGACAGCGAGCCAUGCAGGAACCUCGUUUGGUUUCUGCGUGCUGUCAACAUAAUCUGAACA_CCAC___AACUACGAAACAGAAAG .....((((((((..(((..........))))))(((((((((((......))))))))))))))))......................................... (-24.65 = -24.48 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:51:39 2006