
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,397,575 – 27,397,713 |
| Length | 138 |
| Max. P | 0.988512 |

| Location | 27,397,575 – 27,397,679 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -21.12 |
| Consensus MFE | -13.80 |
| Energy contribution | -13.96 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27397575 104 + 27905053 AGCUUAGCUAUUU-UUCCGCCCAUU--------C-AUAAUCGAUAAUCUAUCGAUAAAUACAAUCGAACCAGUAAGUGAAUCUCAACUAGUUCCU-U--UUCAAUCCAGUGAAG--AAA- ..........(((-((((.......--------.-...(((((((...)))))))..........((((.(((.((.....))..))).))))..-.--.........).))))--)).- ( -12.70) >DroSec_CAF1 13648 116 + 1 AGCUGAAAUAUUC-GUCCGCCCAUUCAACCAUUCAACUAUCGAUACAUUAUCGAUACUAACUAUCGAACCAGUAAGUGAAUCUCAACUGGUUCCUUU--UUUAAUCCACUGCACACAAA- .((.(........-.......................((((((((...)))))))).........((((((((.((.....))..))))))))....--.........).)).......- ( -18.50) >DroSim_CAF1 13702 108 + 1 AGCUUAAAUAUUU-GUCCGCCCAUU--------CGACUAUCGAUACGUUAUCGAUACUAACAAUCGAACCAGUAAGUGAAUCUCAACUGGUUCCGUU--UUUAAUCCACUGCAGACAAA- ..........(((-(((.((.....--------....((((((((...)))))))).(((.(((.((((((((.((.....))..)))))))).)))--.))).......)).))))))- ( -25.10) >DroEre_CAF1 14407 108 + 1 AGUUUAACUAUUCCAGCCGCCUACA--------C-AAAAUCGAUAAGUUAUCGACGCAUAUAAUCGAACCAGUAAGUGAAUUACCGCUGGUUCAUUUUG--UA-UUUACUACAGGCAAAA ..................((((...--------.-....((((((...))))))...((((((..((((((((..(((...))).))))))))...)))--))-).......)))).... ( -23.10) >DroYak_CAF1 13810 107 + 1 AGCUUAACUAU---AUCCGCCUAUU--------C-AUAAUCGAUAAGUUAUCGAUAUGUAUAAUCGAACCAGUAAGUGAACCACCGCUGGUUCAUUUGGUUUA-AUUACUACAGACAAAA ...........---...........--------.-...(((((((...))))))).((((((((..((((((...((((((((....))))))))))))))..-)))).))))....... ( -26.20) >consensus AGCUUAACUAUUC_AUCCGCCCAUU________C_ACAAUCGAUAAGUUAUCGAUACAUACAAUCGAACCAGUAAGUGAAUCUCAACUGGUUCCUUU__UUUAAUCCACUGCAGACAAA_ ......................................(((((((...)))))))..........((((((((............))))))))........................... (-13.80 = -13.96 + 0.16)



| Location | 27,397,575 – 27,397,679 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -17.16 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27397575 104 - 27905053 -UUU--CUUCACUGGAUUGAA--A-AGGAACUAGUUGAGAUUCACUUACUGGUUCGAUUGUAUUUAUCGAUAGAUUAUCGAUUAU-G--------AAUGGGCGGAA-AAAUAGCUAAGCU -...--.((((...((((((.--.-..((((((((.(((.....)))))))))))(((.......))).........)))))).)-)--------))..(((....-.....)))..... ( -19.10) >DroSec_CAF1 13648 116 - 1 -UUUGUGUGCAGUGGAUUAAA--AAAGGAACCAGUUGAGAUUCACUUACUGGUUCGAUAGUUAGUAUCGAUAAUGUAUCGAUAGUUGAAUGGUUGAAUGGGCGGAC-GAAUAUUUCAGCU -(((((.(((..(.(((((..--....((((((((.(((.....))))))))))).....((((((((((((...)))))))).)))).))))).)....))).))-))).......... ( -30.00) >DroSim_CAF1 13702 108 - 1 -UUUGUCUGCAGUGGAUUAAA--AACGGAACCAGUUGAGAUUCACUUACUGGUUCGAUUGUUAGUAUCGAUAACGUAUCGAUAGUCG--------AAUGGGCGGAC-AAAUAUUUAAGCU -(((((((((((((((((...--(((.......)))..))))))))...(.((((((((((..(((.((....)))))..)))))))--------))).)))))))-))).......... ( -35.10) >DroEre_CAF1 14407 108 - 1 UUUUGCCUGUAGUAAA-UA--CAAAAUGAACCAGCGGUAAUUCACUUACUGGUUCGAUUAUAUGCGUCGAUAACUUAUCGAUUUU-G--------UGUAGGCGGCUGGAAUAGUUAAACU .....((.((.((..(-((--((((.((((((((.(((.....)))..)))))))).........(((((((...))))))))))-)--------)))..)).)).))............ ( -26.20) >DroYak_CAF1 13810 107 - 1 UUUUGUCUGUAGUAAU-UAAACCAAAUGAACCAGCGGUGGUUCACUUACUGGUUCGAUUAUACAUAUCGAUAACUUAUCGAUUAU-G--------AAUAGGCGGAU---AUAGUUAAGCU .(((((((((.(((((-..(((((..(((((((....))))))).....)))))..))))).((((((((((...)))))).)))-)--------.))))))))).---........... ( -31.40) >consensus _UUUGUCUGCAGUGGAUUAAA__AAAGGAACCAGUUGAGAUUCACUUACUGGUUCGAUUGUAAGUAUCGAUAACUUAUCGAUUAU_G________AAUGGGCGGAC_AAAUAGUUAAGCU ....((((((.................(((((((..............)))))))..........(((((((...)))))))..................)))))).............. (-17.16 = -17.44 + 0.28)

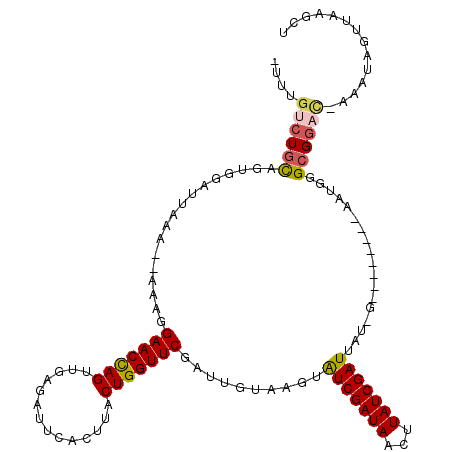

| Location | 27,397,605 – 27,397,713 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.33 |
| Mean single sequence MFE | -17.23 |
| Consensus MFE | -11.56 |
| Energy contribution | -11.52 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27397605 108 + 27905053 CGAUAAUCUAUCGAUAAAUACAAUCGAACCAGUAAGUGAAUCUCAACUAGUUCCU-U--UUCAAUCCAGUGAAG--AAA-CAAGAAAUUUUAAGA-----ACAAAAAA-AACUACCAGAU .....((((.(((((.......)))))...(((.(((........))).((((.(-(--((((......)))))--)..-.((....))....))-----))......-.)))...)))) ( -10.80) >DroSec_CAF1 13687 107 + 1 CGAUACAUUAUCGAUACUAACUAUCGAACCAGUAAGUGAAUCUCAACUGGUUCCUUU--UUUAAUCCACUGCACACAAA-CAAGAAAUGUUAAGA-----AAAGAAAAUAA-----AUAU (...(((((.((((((.....))))((((((((.((.....))..))))))))....--....................-...)))))))...).-----...........-----.... ( -15.40) >DroSim_CAF1 13733 106 + 1 CGAUACGUUAUCGAUACUAACAAUCGAACCAGUAAGUGAAUCUCAACUGGUUCCGUU--UUUAAUCCACUGCAGACAAA-CAAGAAAUCUUAAGA-----AAAGAAAA-AA-----AUAU (((((...)))))....(((.(((.((((((((.((.....))..)))))))).)))--.)))................-.......((((....-----.))))...-..-----.... ( -14.70) >DroEre_CAF1 14438 111 + 1 CGAUAAGUUAUCGACGCAUAUAAUCGAACCAGUAAGUGAAUUACCGCUGGUUCAUUUUG--UA-UUUACUACAGGCAAAACUAAAAAG-CUAAAA----AUUAAUAAA-AACUACAUGAU (((((...)))))..((........((((((((..(((...))).))))))))...(((--((-.....)))))))............-......----.........-........... ( -17.40) >DroYak_CAF1 13838 117 + 1 CGAUAAGUUAUCGAUAUGUAUAAUCGAACCAGUAAGUGAACCACCGCUGGUUCAUUUGGUUUA-AUUACUACAGACAAAACUAAAAAG-CCAAGACAAUAUAUAUAAA-AACUUAAUGGU ((.((((((....((((((((..((((((((((..(((...))).))))))))..(((((((.-.....................)))-))))))..))))))))...-)))))).)).. ( -27.85) >consensus CGAUAAGUUAUCGAUACAUACAAUCGAACCAGUAAGUGAAUCUCAACUGGUUCCUUU__UUUAAUCCACUGCAGACAAA_CAAGAAAU_UUAAGA_____AAAAAAAA_AACU___AGAU (((((...)))))............((((((((............))))))))................................................................... (-11.56 = -11.52 + -0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:51:36 2006