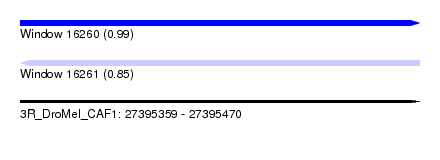
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,395,359 – 27,395,470 |
| Length | 111 |
| Max. P | 0.994082 |

| Location | 27,395,359 – 27,395,470 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -39.54 |
| Consensus MFE | -23.66 |
| Energy contribution | -26.54 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27395359 111 + 27905053 CAUGCUGUGUUCUUUUCCCCUUUUUCCUUUUUUUUAUGGGCUUGGCUUUGGCCUAGCCGCAUCUU---GAAGAGGGAACACGGAGGCUGGGGAAUAGAGGAUGGCUUGCUUGUU ((.(((((.((((.((((((..((((((((((...(((((((.(((....))).)))).)))...---))))))))))(.....)...)))))).)))).))))).))...... ( -43.50) >DroSec_CAF1 11441 106 + 1 CAUGCUGUGUUCUUUUCUCCUUUUUCCUUU-UUUUAUGGGCUUGGCCU-AGCCAAGAUGCGGCU------AGAGGGAACACGGAGGGUGGAGGAGAGAGGAGGGCUUGCUUGUU ((.((.((.(((((((((((((..((((((-.....((..(((...((-((((.......))))------)).)))..)).))))))..))))))))))))).))..)).)).. ( -42.60) >DroSim_CAF1 11488 110 + 1 CAUGCUGUGUUCUUUUCUCCUUUUUCCUUA-UUUUAUGGGCUUGGCUUUGGCCUAGCCGCAUCUU---GAAGGGGGAACACGGAGGGUGGAGGAGAGAGGAGGGCUUGCUUGUU ((.((.((.(((((((((((((.(((((..-(((.(((((((.(((....))).)))).)))...---)))..)))))(((.....)))))))))))))))).))..)).)).. ( -45.90) >DroEre_CAF1 12112 103 + 1 CAUGCUGUGCUCUUUACC----------UU-UUUUAUGGGCUUGGUUUUGGCCCAGCCGCAUCAGGAGGAAGAGGGGAAAUGGAGGCUGAGAAGCAGAGGAUGGCUUGCCGGUU ((.(((((.((((...((----------((-((((.((((((.......)))))).((......))..)))))))).........(((....))))))).))))).))...... ( -32.90) >DroYak_CAF1 11504 102 + 1 CAUGCUGUGUUCCUUACC--------CUUU-UUUUAUGGGCUUGGUUUUGGCCUAGCCGCAUCGU---GAAGAGGGGAUACGGAGGCCGAGAAGAAGUGGAUGGUCUGCCGGUU ................((--------((((-((..(((((((.(((....))).)))).)))...---))))))))....(((((((((............)))))).)))... ( -32.80) >consensus CAUGCUGUGUUCUUUUCCCCUUUUUCCUUU_UUUUAUGGGCUUGGCUUUGGCCUAGCCGCAUCUU___GAAGAGGGAACACGGAGGCUGGGGAAGAGAGGAUGGCUUGCUUGUU ((.(((((.(((((((((((..(((((.........((((((.(((....))).)))).))............(......))))))..))))))))))).))))).))...... (-23.66 = -26.54 + 2.88)

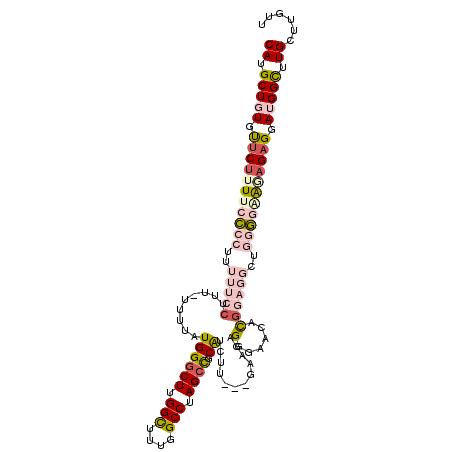
| Location | 27,395,359 – 27,395,470 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -25.71 |
| Consensus MFE | -10.68 |
| Energy contribution | -11.92 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27395359 111 - 27905053 AACAAGCAAGCCAUCCUCUAUUCCCCAGCCUCCGUGUUCCCUCUUC---AAGAUGCGGCUAGGCCAAAGCCAAGCCCAUAAAAAAAAGGAAAAAGGGGAAAAGAACACAGCAUG .........((.....(((.((((((.((....)).((((......---...(((.((((.(((....))).)))))))........))))...)))))).))).....))... ( -30.13) >DroSec_CAF1 11441 106 - 1 AACAAGCAAGCCCUCCUCUCUCCUCCACCCUCCGUGUUCCCUCU------AGCCGCAUCUUGGCU-AGGCCAAGCCCAUAAAA-AAAGGAAAAAGGAGAAAAGAACACAGCAUG ............(((((.(.((((.(((.....))).....(((------(((((.....)))))-)))..............-..)))).).)))))................ ( -21.30) >DroSim_CAF1 11488 110 - 1 AACAAGCAAGCCCUCCUCUCUCCUCCACCCUCCGUGUUCCCCCUUC---AAGAUGCGGCUAGGCCAAAGCCAAGCCCAUAAAA-UAAGGAAAAAGGAGAAAAGAACACAGCAUG .....((......((...((((((.(((.....))).....((((.---...(((.((((.(((....))).)))))))....-.))))....))))))...)).....))... ( -26.30) >DroEre_CAF1 12112 103 - 1 AACCGGCAAGCCAUCCUCUGCUUCUCAGCCUCCAUUUCCCCUCUUCCUCCUGAUGCGGCUGGGCCAAAACCAAGCCCAUAAAA-AA----------GGUAAAGAGCACAGCAUG ....(((((((........))))....)))..........(((((...(((.(((.((((((.......)).)))))))....-.)----------))..)))))......... ( -23.40) >DroYak_CAF1 11504 102 - 1 AACCGGCAGACCAUCCACUUCUUCUCGGCCUCCGUAUCCCCUCUUC---ACGAUGCGGCUAGGCCAAAACCAAGCCCAUAAAA-AAAG--------GGUAAGGAACACAGCAUG .....((...................((((((((((((........---..)))))))..)))))....((..((((......-...)--------)))..))......))... ( -27.40) >consensus AACAAGCAAGCCAUCCUCUCUUCCCCAGCCUCCGUGUUCCCUCUUC___AAGAUGCGGCUAGGCCAAAGCCAAGCCCAUAAAA_AAAGGAAAAAGGGGAAAAGAACACAGCAUG .........((.....(((...((((.......(((((.............)))))((((.(((....))).))))..................))))...))).....))... (-10.68 = -11.92 + 1.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:51:31 2006