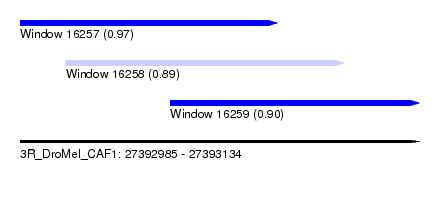
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,392,985 – 27,393,134 |
| Length | 149 |
| Max. P | 0.974563 |

| Location | 27,392,985 – 27,393,081 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -21.25 |
| Energy contribution | -22.17 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27392985 96 + 27905053 GCAAGCC-----------------------UAGAGAACGUGGCCAAGUGUGG-GCUUAUUGGCGUUGACAGCUAUGCAAAUGCUUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUU ..(((((-----------------------..(..(((((((((((((((..-((....((((.......)))).))..))))))))))))..((((...)))))))..)...))))).. ( -35.70) >DroPse_CAF1 49813 87 + 1 ------------------------------UCCAGAACGUGGCCAAGUGUGG-GC--AUUGCCGGUGACAGCUAUGCAAAUGCCUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUU ------------------------------........(((((((.((((..-((--((.((.(....).)).))))..)))).))))))).(((((((((........))))))))).. ( -34.60) >DroSec_CAF1 9100 97 + 1 GCUAGCC-----------------------UAGAGAACGUGGCCAAGUGUGGGGCUUAUUGGUGUUGACAACUAUGCAAAUGCUUGACCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUU ((((((.-----------------------........((((.(((((((...((....((((.......)))).))..))))))).))))..((((...)))))))))).......... ( -25.10) >DroWil_CAF1 11802 76 + 1 -----------------------------------------GCCUAGUGGGG-GC--AUUGGCGUUGACAGCUAUGCAAAUGCCUGGCCAUUAAGUUGUUGACUGUUAGCAAUGGCUUUU -----------------------------------------(((((((((((-((--(((.((((........)))).)))))))..)))))).((((((((...))))))))))).... ( -27.60) >DroAna_CAF1 9864 117 + 1 ACGUGGCUGGGGGAUGGGGAAUGGGGGGCACAAAGAACGUGGCCAAGUGUGG-GC--AUUGGCGUUGACAGCUAUGCAAAUGCUUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUU ..(((.((.................)).))).......((((((((((((..-((--((.(((.......)))))))..)))))))))))).(((((((((........))))))))).. ( -39.73) >DroPer_CAF1 48506 87 + 1 ------------------------------UCCAGAACGUGGCCAAGUGUGG-GC--AUUGCCGGUGACAGCUAUGCAAAUGCCUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUU ------------------------------........(((((((.((((..-((--((.((.(....).)).))))..)))).))))))).(((((((((........))))))))).. ( -34.60) >consensus ______________________________UACAGAACGUGGCCAAGUGUGG_GC__AUUGGCGUUGACAGCUAUGCAAAUGCCUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUU ......................................(((((((.((((...((....((((.......)))).))..)))).))))))).(((((((((........))))))))).. (-21.25 = -22.17 + 0.92)

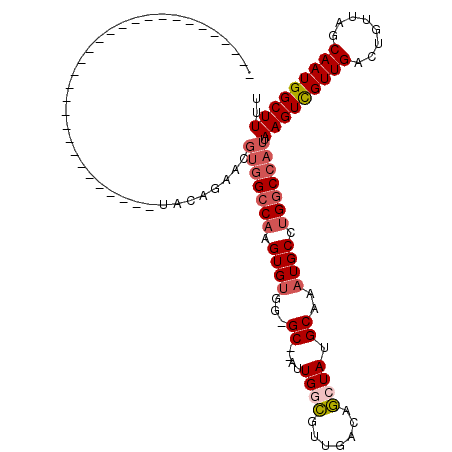
| Location | 27,393,002 – 27,393,106 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -43.43 |
| Consensus MFE | -27.62 |
| Energy contribution | -28.73 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27393002 104 + 27905053 GGCCAAGUGUGG-GCUUAUUGGCGUUGACAGCUAUGCAAAUGCUUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCUUGACUUUGGCCCG---------------G ((((((((((..-((....((((.......)))).))..)))))))))).((((((((((((.((..(((....)))..)).))))))))))))..........---------------. ( -41.20) >DroPse_CAF1 49823 115 + 1 GGCCAAGUGUGG-GC--AUUGCCGGUGACAGCUAUGCAAAUGCCUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCUUGACUUUGGCCUC-GACACA-GGCCUCAA (((((.((((..-((--((.((.(....).)).))))..)))).))))).((((((((((((.((..(((....)))..)).))))))))))))....(((((.-.....)-)))).... ( -49.80) >DroSec_CAF1 9117 105 + 1 GGCCAAGUGUGGGGCUUAUUGGUGUUGACAACUAUGCAAAUGCUUGACCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCUUGACUUUGGCCCG---------------G (((((((.((.((((....((((.......)))).......)))).))).((((((((((((.((..(((....)))..)).))))))))))))..))))))..---------------. ( -34.30) >DroWil_CAF1 11802 111 + 1 -GCCUAGUGGGG-GC--AUUGGCGUUGACAGCUAUGCAAAUGCCUGGCCAUUAAGUUGUUGACUGUUAGCAAUGGCUUUUAAUUAGCAGCU-----UUGGCCUAAGGCACAAGGCCAAAA -((((.(((.((-((--(((.((((........)))).)))))))(((((..((((((((((.((..(((....)))..)).)))))))))-----)))))).....))).))))..... ( -46.40) >DroAna_CAF1 9904 101 + 1 GGCCAAGUGUGG-GC--AUUGGCGUUGACAGCUAUGCAAAUGCUUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCUUGACUU-AGCCCG---------------G ((((((((((..-((--((.(((.......)))))))..)))))))))).((((((((((((.((..(((....)))..)).))))))))))))...-......---------------. ( -43.20) >DroPer_CAF1 48516 115 + 1 GGCCAAGUGUGG-GC--AUUGCCGGUGACAGCUAUGCAAAUGCCUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCCUGACUUUGGCCUC-GACACA-GGCCUCAA (((((.((((..-((--((.((.(....).)).))))..)))).))))).(((.((((((((.((..(((....)))..)).)))))))).)))....(((((.-.....)-)))).... ( -45.70) >consensus GGCCAAGUGUGG_GC__AUUGGCGUUGACAGCUAUGCAAAUGCCUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCUUGACUUUGGCCCG_______________A (((((.((((...((....((((.......)))).))..)))).))))).((((((((((((.((..(((....)))..)).)))))))))))).......................... (-27.62 = -28.73 + 1.11)

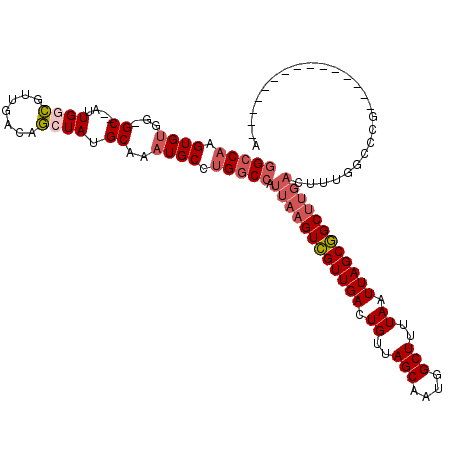
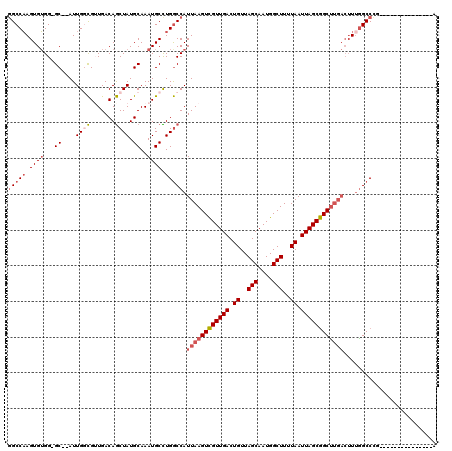
| Location | 27,393,041 – 27,393,134 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.15 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -22.55 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27393041 93 + 27905053 UGCUUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCUUGACUUUGGCCCG-------------GCCCGGGAUUGUGAUGGUCUAGAGCCCGA .(((.(((((((((((((((((.((..(((....)))..)).))))))))))))...))))).)-------------)).((((.((.((......)).)))))). ( -36.20) >DroPse_CAF1 49860 105 + 1 UGCCUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCUUGACUUUGGCCUCGACACAGGCCUCAACCU-GGAUCGUUCUGAACCCCAGCAUGG (((..(((((((((((((((((.((..(((....)))..)).))))))))))))...))))).(((..((((......)))-)..)))............)))... ( -36.80) >DroSim_CAF1 9165 93 + 1 UGCUUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCUUGACUUUGGCCCG-------------GCCCGGGAUGGGGAUGGUCCACAGCACGG .(((.(((((((((((((((((.((..(((....)))..)).))))))))))))...))))).)-------------))(((...((.((.....)).))...))) ( -37.00) >DroYak_CAF1 9241 92 + 1 UGCUUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCUUGACUUUGCCCCG-------------GCCU-GGAUUGAGAUGGCCCAGAGCCCGG .(((((((((((((((((((((.((..(((....)))..)).))))))))))..((..((....-------------))..-)).....)))))))..)))).... ( -32.90) >DroAna_CAF1 9941 85 + 1 UGCUUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCUUGACUU-AGCCCG-------------GCCU-GGAUC------GUUUUGAACUUGG .(((.(((..((((((((((((.((..(((....)))..)).))))))))))))...-.))).)-------------))..-.....------............. ( -25.10) >DroPer_CAF1 48553 105 + 1 UGCCUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCCUGACUUUGGCCUCGACACAGGCCUCAACCU-GGAUCGUUCUGAACCCCAGCAUGG (((..((((((((.((((((((.((..(((....)))..)).)))))))).)))...))))).(((..((((......)))-)..)))............)))... ( -32.70) >consensus UGCUUGGCCAUUAAGUCGUUGACUGUUAGCAAUGGCUUUUAAUUAGCGGCUUGACUUUGGCCCG_____________GCCU_GGAUCGUGAUGGUCCACAGCACGG .....(((((((((((((((((.((..(((....)))..)).))))))))))))...)))))............................................ (-22.55 = -23.22 + 0.67)

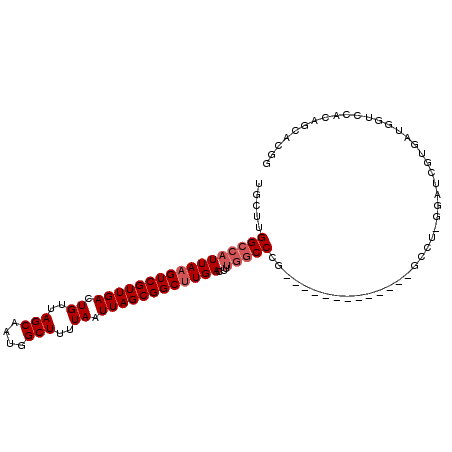
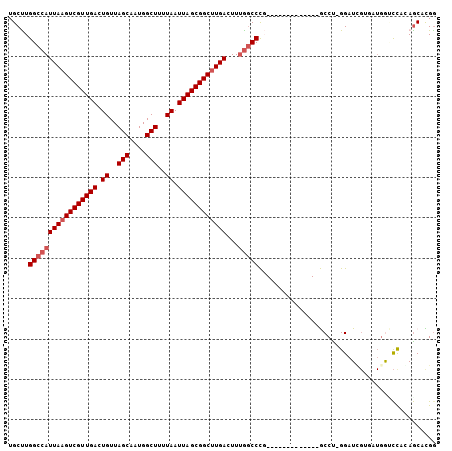
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:51:29 2006