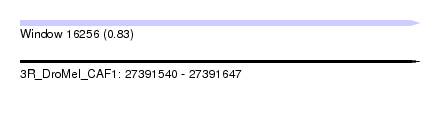
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,391,540 – 27,391,647 |
| Length | 107 |
| Max. P | 0.830849 |

| Location | 27,391,540 – 27,391,647 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.85 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -20.68 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27391540 107 + 27905053 AACUAUAU----UGCUGCUCUCAAACCGCAUUAACCCGCAAAGUUUGCCAAAUAAUUUUGUAUCAAAAUGUUUUGCGCCAGGCUUCUGUGCAGGGCGGCAUUUCCAAAUUU ........----(((((((((((((..(((((.....((((((((........)))))))).....))))))))).(((((....))).)))))))))))........... ( -26.20) >DroPse_CAF1 48029 104 + 1 AACUAUUUAUUCAGCUGCUAAC-------AUUUUCACACAAAGUUUGCCAAAUAAUUUUGUAUCAAAAUGUUUUGCGCCAGGCUUCUGUGCUAGGCGGCGUUUCCCAGCUC ............(((((..(((-------(((((.(.((((((((........)))))))).).))))))))...((((.(((......))).))))........))))). ( -26.90) >DroEre_CAF1 8309 101 + 1 AACUAUAU----UGCUGCUCUCAA------UUACCCCGCAAAGUUUGCCAAAUAAUUUUGUAUCAAAAUGUUUUGCGCCAGGCUUCUGUGCAGGGCGGCAUUUCCAAAUUU ........----(((((((((...------......((((((((((((((((..((((((...))))))..)))).).))))))).)))).)))))))))........... ( -24.80) >DroYak_CAF1 7670 101 + 1 AACUAUAU----UGCUGUCCUCAA------UUAACCCGCAAAGUUUGCCAAAUAAUUUUGUAUCAAAAUGUUUUGCGCCAGGCUUCUGUGCAGGGCGGCAUUUCCAAAUUU ........----(((((((((...------......((((((((((((((((..((((((...))))))..)))).).))))))).)))).)))))))))........... ( -25.10) >DroAna_CAF1 8314 100 + 1 ACCUAUUA----AGCUGCUGAC-------AUUUUCCCACAAAGUUUGCCAAAUAAUUUUGUAUCAAAAUGUUUUGCGCCAGGCUUCUGUGCAAGGCGGCAUUUCUAAAUUU ........----.(((((((((-------(((((...((((((((........))))))))...))))))))((((((.((....))))))))))))))............ ( -27.00) >DroPer_CAF1 46690 104 + 1 AACUAUUUAUUCAGCUGCUAAC-------AUUUUCACACAAAGUUUGCCAAAUAAUUUUGUAUCAAAAUGUUUUGCGCCAGGCUUCUGUGCUAGGCGGCGUUUCCCAGCUC ............(((((..(((-------(((((.(.((((((((........)))))))).).))))))))...((((.(((......))).))))........))))). ( -26.90) >consensus AACUAUAU____AGCUGCUCAC_______AUUAUCCCACAAAGUUUGCCAAAUAAUUUUGUAUCAAAAUGUUUUGCGCCAGGCUUCUGUGCAAGGCGGCAUUUCCAAAUUU .............((((((..................((((((((........))))))))...........((((((.((....))))))))))))))............ (-20.68 = -20.23 + -0.44)

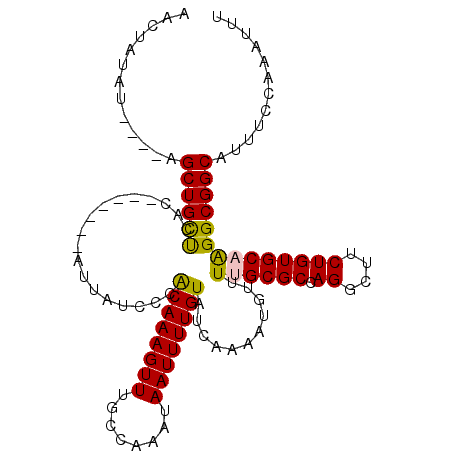
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:51:27 2006