| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,208,966 – 3,209,096 |
| Length | 130 |
| Max. P | 0.700186 |

| Location | 3,208,966 – 3,209,058 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.71 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.33 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
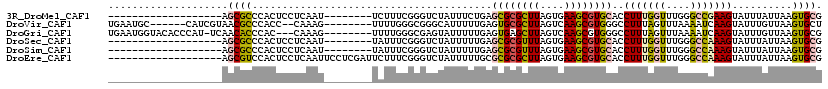
>3R_DroMel_CAF1 3208966 92 + 27905053 -------------------AGCGCCCACUCCUCAAU--------UCUUUCGGGUCUAUUUCUGAGCGCGCUUAGUGAAGCGUGCACCUUUGGUUUGGGCCGAAGUAUUUAUUAAGUGCG -------------------.(.(((((..((.....--------....(((((......)))))((((((((....))))))))......))..))))))...(((((.....))))). ( -31.30) >DroVir_CAF1 36712 103 + 1 UGAAUGC------CAUCGUAACGCCCACC--CAAAG--------UUUUGGGCGGGCAUUUUUGAGUGCGCUUAGUCAAGCGUGGGCCUUUAGUUUAAAAUCAAGUAUUUGUUAAGUGCU ((((.((------(........((((.((--(((..--------..))))).))))..........((((((....)))))).))).))))...........((((((.....)))))) ( -31.40) >DroGri_CAF1 29996 107 + 1 UGAAUGGUACACCCAU-UCAACACCCAC---CAAAG--------UUUUGGGCGAGUAUUUUUGAGUGAGCUUAGUCAAGCGUGGGCCUUUAGUUUAAAAUCAAGUAUUUGUUAAGUGCG (((((((.....))))-))).(((...(---(((..--------..))))((((((((..((((.((((((..(((.......)))....))))))...))))))))))))...))).. ( -25.60) >DroSec_CAF1 28312 92 + 1 -------------------AGCGCCCACUCCUCAAU--------UAUUUCGGGUCUAUUUUUGAGCGCGUUUAGUGAAGCGUGCACCUUUGGUUUGGGCCAAAGUAUUUAUUAAGUGCG -------------------.(.(((((..((.....--------....(((((......)))))((((((((....))))))))......))..))))))...(((((.....))))). ( -26.30) >DroSim_CAF1 35134 92 + 1 -------------------AGCGCCCACUCCUCAAU--------UAUUUCGGGUCUAUUUUUGAGCGCGUUUAGUGAAGCGUGCACCUUUGGUUUGGGCCAAAGUAUUUAUUAAGUGCG -------------------.(.(((((..((.....--------....(((((......)))))((((((((....))))))))......))..))))))...(((((.....))))). ( -26.30) >DroEre_CAF1 27630 100 + 1 -------------------AGCGUCCACUCCUCAAUUCCUCGAUUCUUUCGGGUCUAUUUUUGCGCGCGCUUAGUGAAGCGUGCACCUUUGGUUUGGGCCAAAGUAUUUAUUAAGUGCG -------------------.(.(((((..((.......(((((.....)))))...........((((((((....))))))))......))..))))))...(((((.....))))). ( -29.90) >consensus ___________________AGCGCCCACUCCUCAAU________UAUUUCGGGUCUAUUUUUGAGCGCGCUUAGUGAAGCGUGCACCUUUGGUUUGGGCCAAAGUAUUUAUUAAGUGCG ....................((((........................................((((((((....))))))))..(((((((....)))))))..........)))). (-16.30 = -16.33 + 0.03)
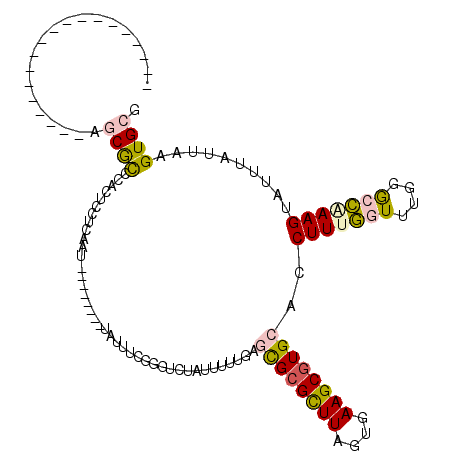
| Location | 3,208,983 – 3,209,096 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.95 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -16.46 |
| Energy contribution | -15.97 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3208983 113 + 27905053 ----UCUUUCGGGUCUAUUUCUGAGCGCGCUUAGUGAAGCGUGCACCUUUGGUUUGGGCCGAAGUAUUUAUUAAGUGCGGAAUGUGGGUCGCAUUACGGAGGACGUCGAGAUGCCGA-- ----....((((..(((((((((.((((((((....))))))))..(((((((....))))))).............))))).))))..)(((((.(((......))).))))))))-- ( -39.20) >DroVir_CAF1 36740 103 + 1 ----UUUUGGGCGGGCAUUUUUGAGUGCGCUUAGUCAAGCGUGGGCCUUUAGUUUAAAAUCAAGUAUUUGUUAAGUGCUGCGAUUCGAUUUUAUGAC---UCG--CCAUG-------AA ----.....(((((((((..((((((((((((((.((((..(..(..((((...))))..)..)..))))))))))))....))))))....))).)---)))--))...-------.. ( -26.70) >DroGri_CAF1 30028 103 + 1 ----UUUUGGGCGAGUAUUUUUGAGUGAGCUUAGUCAAGCGUGGGCCUUUAGUUUAAAAUCAAGUAUUUGUUAAGUGCGGAGAUUCGAUUUUAUGGG---CCA--CCAUG-------AA ----...((((((.......(((((....))))).....))).(((((......(((((((.(((.(((((.....))))).))).))))))).)))---)).--)))..-------.. ( -24.10) >DroSec_CAF1 28329 113 + 1 ----UAUUUCGGGUCUAUUUUUGAGCGCGUUUAGUGAAGCGUGCACCUUUGGUUUGGGCCAAAGUAUUUAUUAAGUGCGGAAUUUGGGUCGCAUUACGGAGGACGCCGAGAUGCCGA-- ----((((((((((((...((((.((((((((....))))))))..(((((((....))))))).........(((((((........))))))).)))))))).))))))))....-- ( -38.80) >DroSim_CAF1 35151 113 + 1 ----UAUUUCGGGUCUAUUUUUGAGCGCGUUUAGUGAAGCGUGCACCUUUGGUUUGGGCCAAAGUAUUUAUUAAGUGCGGAAUGUGGGUCGCAUUACGGAGGACGUCGAGAUGCCGA-- ----((((((((((((...((((.((((((((....))))))))..(((((((....))))))).........(((((((........))))))).)))))))).))))))))....-- ( -35.80) >DroEre_CAF1 27651 93 + 1 CGAUUCUUUCGGGUCUAUUUUUGCGCGCGCUUAGUGAAGCGUGCACCUUUGGUUUGGGCCAAAGUAUUUAUUAAGUGCGGAAUCUGGGUCGCA-------------------------- (((.....)))(..(((.((((((((((((((....))))))))..(((((((....)))))))............))))))..)))..)...-------------------------- ( -32.70) >consensus ____UAUUUCGGGUCUAUUUUUGAGCGCGCUUAGUGAAGCGUGCACCUUUGGUUUGGGCCAAAGUAUUUAUUAAGUGCGGAAUUUGGGUCGCAUUAC___GGA__CCGAG_________ .........(((.((...(((((.((((((((....))))))))..(((((((....))))))).............)))))...)).)))............................ (-16.46 = -15.97 + -0.49)

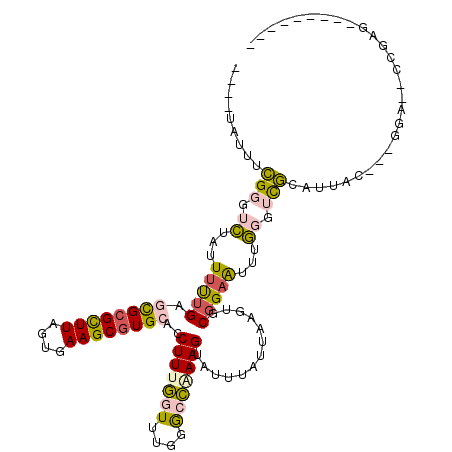
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:30 2006