
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,363,702 – 27,363,812 |
| Length | 110 |
| Max. P | 0.875984 |

| Location | 27,363,702 – 27,363,812 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.65 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
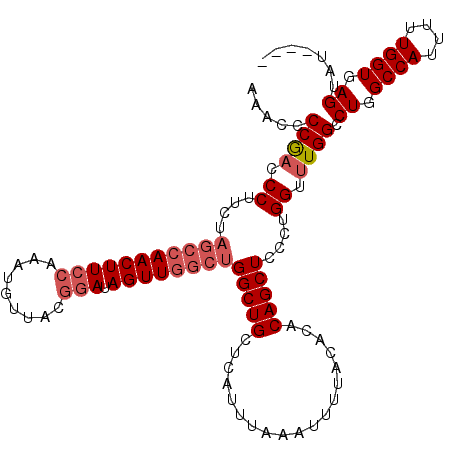
>3R_DroMel_CAF1 27363702 110 + 27905053 ----AUACUCACCAAAAAUGGCCAGGCCAAACCAGGGAGCUGUGUGUAAAAUUUAAAUGAGCAGCCAGCCAACUAUCCGUAAAAUUUGGAAGUUGGCUAGAAGGGUCGGGGUUU ----......(((.....(((((.(((...(((((....))).))((.............)).)))((((((((.((((.......)))))))))))).....))))).))).. ( -31.32) >DroSec_CAF1 109219 110 + 1 ----AUACUCACCAAAAAUGGCCAGGCCAAACCAGGGAGCUGUGUGUAAAAUUUAAAUGAGCAGCCAGACAACUAUCCGUAACUUUUGGAAGUUGGCUAGAAGGGUCGGGGAUU ----...((((((.....(((((((.(((((..(.((((((((.................))))).((....)).))).)....)))))...)))))))....))).))).... ( -26.43) >DroSim_CAF1 73400 110 + 1 ----AUACUCACCAAAAAUGGCCAGGCCAAACCAGGGAGCUGUGUGUAAAAUUUAAAUGAGCAGCCAGCCAACUAUCCGUAACAUUUGGAAGUUGGCUAGAAGGGUCGGGGAUU ----...((((((......(((...((((.(((((....))).))............)).)).)))((((((((.((((.......)))))))))))).....))).))).... ( -31.22) >DroEre_CAF1 73714 110 + 1 ----AUACUCACCAAAAAUGGCCAGGCCAAACCAGGGAGCUGUGUGUAAAAUUUAAAUGAGCAGCCAGCCAACUAUCCGUAACAUUUGGAAGUUGGCUAGAAGGGUCGGGGUUU ----......(((.....(((((.(((...(((((....))).))((.............)).)))((((((((.((((.......)))))))))))).....))))).))).. ( -31.32) >DroYak_CAF1 66584 110 + 1 ----AUACUCACCAAAAAUGGCCAGGCCAAACCAGGGAGCUGUGUGUAAAAUUUAAAUGAGCAGCCAGCCAACUAUCCGUAACAUUUGGAAGUUGGCUAGAAGGGUCGGGGUUU ----......(((.....(((((.(((...(((((....))).))((.............)).)))((((((((.((((.......)))))))))))).....))))).))).. ( -31.32) >DroAna_CAF1 78271 99 + 1 AAAGAGACUCACCAAAAAUGGCCAGGCCAAACCUGGGAGCUGUGUGUAAAAUUUAAAUGAGCAGCCAGCUAACUAUCC-----------AAGUUGGUG---GGGGAUGGGGUU- .......((((((((...((((((((.....))))).(((((..(((.............)))..)))))......))-----------)..))))))---))..........- ( -30.32) >consensus ____AUACUCACCAAAAAUGGCCAGGCCAAACCAGGGAGCUGUGUGUAAAAUUUAAAUGAGCAGCCAGCCAACUAUCCGUAACAUUUGGAAGUUGGCUAGAAGGGUCGGGGUUU .......(((.(((....)))...((((........(.(((((.................))))))((((((((.((((.......)))))))))))).....))))))).... (-26.19 = -26.65 + 0.46)



| Location | 27,363,702 – 27,363,812 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -24.67 |
| Energy contribution | -25.69 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27363702 110 - 27905053 AAACCCCGACCCUUCUAGCCAACUUCCAAAUUUUACGGAUAGUUGGCUGGCUGCUCAUUUAAAUUUUACACACAGCUCCCUGGUUUGGCCUGGCCAUUUUUGGUGAGUAU---- ....((.((((....((((((((((((.........))).)))))))))((((...................)))).....)))).)).((.((((....)))).))...---- ( -30.81) >DroSec_CAF1 109219 110 - 1 AAUCCCCGACCCUUCUAGCCAACUUCCAAAAGUUACGGAUAGUUGUCUGGCUGCUCAUUUAAAUUUUACACACAGCUCCCUGGUUUGGCCUGGCCAUUUUUGGUGAGUAU---- ....((.((((....(((.((((((((.........))).))))).)))((((...................)))).....)))).)).((.((((....)))).))...---- ( -23.71) >DroSim_CAF1 73400 110 - 1 AAUCCCCGACCCUUCUAGCCAACUUCCAAAUGUUACGGAUAGUUGGCUGGCUGCUCAUUUAAAUUUUACACACAGCUCCCUGGUUUGGCCUGGCCAUUUUUGGUGAGUAU---- ....((.((((....((((((((((((.........))).)))))))))((((...................)))).....)))).)).((.((((....)))).))...---- ( -30.81) >DroEre_CAF1 73714 110 - 1 AAACCCCGACCCUUCUAGCCAACUUCCAAAUGUUACGGAUAGUUGGCUGGCUGCUCAUUUAAAUUUUACACACAGCUCCCUGGUUUGGCCUGGCCAUUUUUGGUGAGUAU---- ....((.((((....((((((((((((.........))).)))))))))((((...................)))).....)))).)).((.((((....)))).))...---- ( -30.81) >DroYak_CAF1 66584 110 - 1 AAACCCCGACCCUUCUAGCCAACUUCCAAAUGUUACGGAUAGUUGGCUGGCUGCUCAUUUAAAUUUUACACACAGCUCCCUGGUUUGGCCUGGCCAUUUUUGGUGAGUAU---- ....((.((((....((((((((((((.........))).)))))))))((((...................)))).....)))).)).((.((((....)))).))...---- ( -30.81) >DroAna_CAF1 78271 99 - 1 -AACCCCAUCCCC---CACCAACUU-----------GGAUAGUUAGCUGGCUGCUCAUUUAAAUUUUACACACAGCUCCCAGGUUUGGCCUGGCCAUUUUUGGUGAGUCUCUUU -............---((((((..(-----------((......(((((..((...............))..))))).(((((.....))))))))...))))))......... ( -24.16) >consensus AAACCCCGACCCUUCUAGCCAACUUCCAAAUGUUACGGAUAGUUGGCUGGCUGCUCAUUUAAAUUUUACACACAGCUCCCUGGUUUGGCCUGGCCAUUUUUGGUGAGUAU____ .....((((.((....(((((((((((.........))).))))))))(((((...................)))))....)).)))).((.((((....)))).))....... (-24.67 = -25.69 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:51:13 2006