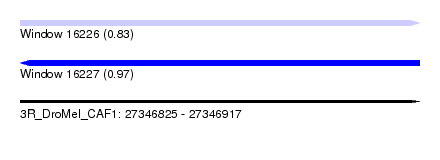
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,346,825 – 27,346,917 |
| Length | 92 |
| Max. P | 0.970216 |

| Location | 27,346,825 – 27,346,917 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -23.49 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27346825 92 + 27905053 AUUUUUGGCAGUCGACGG-------CCGCCGGCUUCUCAAUCAGACUGACCCAGAUAUGGGUCAAGCCAUUGACUUCUGCCA-AGUGCUACCUUU-UUGCC ...((((((((((((.((-------(........(((.....))).(((((((....))))))).))).)))))...)))))-))..........-..... ( -28.80) >DroPse_CAF1 44416 84 + 1 AUUUUUGGCAGUCAACGG-------CCGCUGGCUUCCCAAUCAUUUUGACCCACUUCUGGGCCAAGCCAUUGACUUCUGCCCGUGUCC---------GCA- ......(((((((((.((-------(...(((....)))......(((.((((....)))).)))))).)))))...)))).((....---------)).- ( -22.90) >DroSim_CAF1 56093 85 + 1 AUUUUUGGCAGUCGACGG-------CCGCCGGCUGCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCAUAGUGCUACC--------- .....((((((..(.(((-------((...))))).(((((..(..((((((......))))))..).))))))..))))))..........--------- ( -28.00) >DroEre_CAF1 56948 93 + 1 AUUUUUGGCAGUCGACGG-------CCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCAUUGUGCUACCUU-CUGCCC ......(((((..(..((-------(((..(((...(((((..(..((((((......))))))..).))))).....)))..)).)))..)..-))))). ( -29.80) >DroYak_CAF1 49472 101 + 1 AUUUUUGGCAGUCGACGGCCACCGGCCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUUACUUCUGCCAUUGUGCUACCUUUUUGCCC .....((((((..(.(((((...))))).)(((((((.....))).((((((......))))))))))........))))))................... ( -33.10) >DroAna_CAF1 58719 91 + 1 AUUUUUGGCAGUCGACGG-------CCGCCGGCUCCUCAAUCAGUCUGACCUAGUUUCGGGACAAGCCAUUGACUUGUGC--UUCUGCUGGUUU-CCGCAC ......((.(((((.(..-------..).))))))).......((..((((.(((...((.(((((.......))))).)--)...))))))).-..)).. ( -21.60) >consensus AUUUUUGGCAGUCGACGG_______CCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCAUUGUGCUACCUU__UGCCC .....(((((((((.((((.....)))).)))))............((((((......)))))).))))................................ (-23.49 = -23.62 + 0.13)

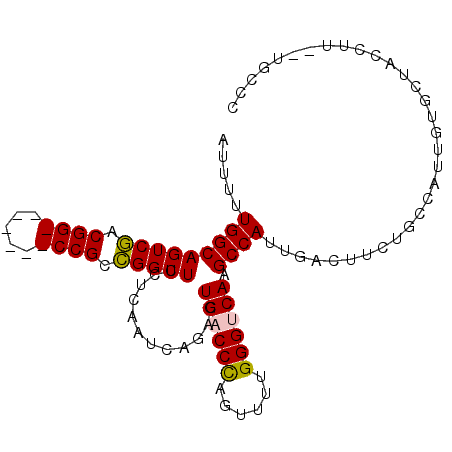

| Location | 27,346,825 – 27,346,917 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -29.28 |
| Energy contribution | -29.68 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27346825 92 - 27905053 GGCAA-AAAGGUAGCACU-UGGCAGAAGUCAAUGGCUUGACCCAUAUCUGGGUCAGUCUGAUUGAGAAGCCGGCGG-------CCGUCGACUGCCAAAAAU ((((.-...((..((..(-((((...(((((..((((.((((((....))))))))))))))).....)))))..)-------)..))...))))...... ( -33.20) >DroPse_CAF1 44416 84 - 1 -UGC---------GGACACGGGCAGAAGUCAAUGGCUUGGCCCAGAAGUGGGUCAAAAUGAUUGGGAAGCCAGCGG-------CCGUUGACUGCCAAAAAU -..(---------(....))((((...(((((((((((((((((....))))))))..((.((((....)))))))-------))))))))))))...... ( -35.90) >DroSim_CAF1 56093 85 - 1 ---------GGUAGCACUAUGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGCAGCCGGCGG-------CCGUCGACUGCCAAAAAU ---------((((((.....(((.(.(((((..((((.((((((....)))))))))))))))...).)))(((..-------..)))).)))))...... ( -31.90) >DroEre_CAF1 56948 93 - 1 GGGCAG-AAGGUAGCACAAUGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGG-------CCGUCGACUGCCAAAAAU .(((((-..((..((....((((...(((((..((((.((((((....))))))))))))))).....))))...)-------)..))..)))))...... ( -38.00) >DroYak_CAF1 49472 101 - 1 GGGCAAAAAGGUAGCACAAUGGCAGAAGUAAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGGCCGGUGGCCGUCGACUGCCAAAAAU ..((.........))....((((((........(((((((((((....))))))).(((.....)))))))(((((((...)))))))..))))))..... ( -40.20) >DroAna_CAF1 58719 91 - 1 GUGCGG-AAACCAGCAGAA--GCACAAGUCAAUGGCUUGUCCCGAAACUAGGUCAGACUGAUUGAGGAGCCGGCGG-------CCGUCGACUGCCAAAAAU (((((.-...)(....)..--)))).((((.((((((.(.((.(...((..(((.....)))..))...).)))))-------)))).))))......... ( -24.30) >consensus GGGCA__AAGGUAGCACAAUGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGG_______CCGUCGACUGCCAAAAAU ....................(((((...(((((((.((((((((....)))))))).)).))))).....(((((((.....))))))).)))))...... (-29.28 = -29.68 + 0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:51:02 2006