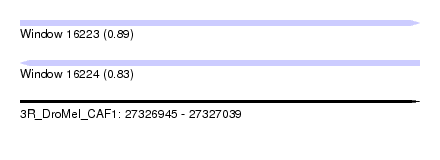
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,326,945 – 27,327,039 |
| Length | 94 |
| Max. P | 0.889667 |

| Location | 27,326,945 – 27,327,039 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.72 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -26.02 |
| Energy contribution | -26.35 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
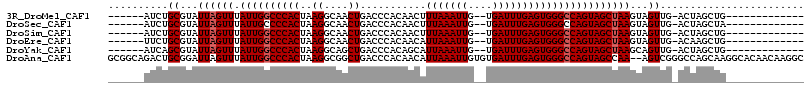
>3R_DroMel_CAF1 27326945 94 + 27905053 ------AUCUGCGUAUUAGUUUAUUGGCCCACUAAGGCAACUGACCCACAACUUUAAAUUG--UGAUUUGAGUGGGCCAGUAGCUAAGUAGUUG-ACUAGCUG------------- ------.((.((...((((((.(((((((((((.((....))....(((((.......)))--)).....)))))))))))))))))...)).)-).......------------- ( -33.10) >DroSec_CAF1 32308 94 + 1 ------AUCUGCGUAUUAGUUUAUUGCCCCACUAAGGCAACUGACCCACAACUUUAAAUUG--UGAUUUGAGUGGGCCAGUAGCUAAGUAGUUG-ACUAGCUA------------- ------.((.((...((((((.((((.((((((.((....))....(((((.......)))--)).....)))))).))))))))))...)).)-).......------------- ( -26.00) >DroSim_CAF1 30217 94 + 1 ------AUCUGCGUAUUAGUUUAUUGGCCCACUAAGGCAACUGACCCACAACUUUAAAUUG--UGAUUUGAGUGGGCCAGUAGCUAAGUAGUUG-ACUAGCUG------------- ------.((.((...((((((.(((((((((((.((....))....(((((.......)))--)).....)))))))))))))))))...)).)-).......------------- ( -33.10) >DroEre_CAF1 36738 94 + 1 ------UUCUGCGUAUUAGUUUAUUGGCCCACUAAGGCAACUGACCCACAACAUUAAAUUG--UGAUUUGAGUGGGCCAGUAGCUAAGUAGUUG-ACAAGCUG------------- ------.((.((...((((((.(((((((((((.((....))....(((((.......)))--)).....)))))))))))))))))...)).)-).......------------- ( -33.00) >DroYak_CAF1 28942 94 + 1 ------AUCAGCGUAUUAGUUUAUUGGCCCACUAAGGCAGCUGACCCACAGCAUUAAAUUG--UGAUUUGAGUGGGCCAGUAGCUAAGCAGUUG-ACUAGCUG------------- ------.(((((...((((((.((((((((((.......((((.....)))).((((((..--..))))))))))))))))))))))...))))-).......------------- ( -36.50) >DroAna_CAF1 36927 114 + 1 GCGGCAGACUGCGGAUUAGUUUAUUGGCCCACUAAGGCGGCUGACCCACAACAUUAAAUUGUGUGAUUUGAGUGGGCCAGUAGCCAA--AGUCGGGCCAGCAAGGCACAACAAGGC ((.((((((((.....))))))..(((((((((..(((.((((.(((((....(((((((....)))))))))))).)))).)))..--))).))))))))...)).......... ( -43.30) >consensus ______AUCUGCGUAUUAGUUUAUUGGCCCACUAAGGCAACUGACCCACAACAUUAAAUUG__UGAUUUGAGUGGGCCAGUAGCUAAGUAGUUG_ACUAGCUG_____________ ..........((...((((((.((((((((((..((....))...........(((((((....)))))))))))))))))))))))...))........................ (-26.02 = -26.35 + 0.33)
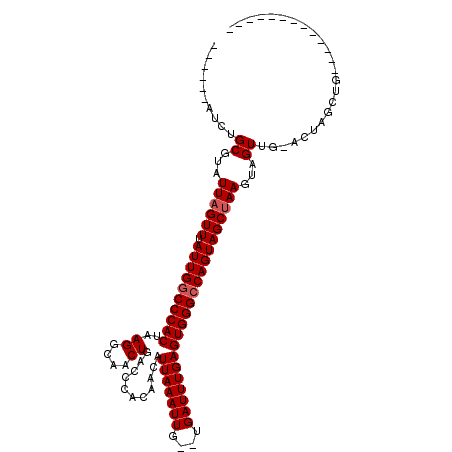

| Location | 27,326,945 – 27,327,039 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 85.72 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -22.81 |
| Energy contribution | -23.37 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27326945 94 - 27905053 -------------CAGCUAGU-CAACUACUUAGCUACUGGCCCACUCAAAUCA--CAAUUUAAAGUUGUGGGUCAGUUGCCUUAGUGGGCCAAUAAACUAAUACGCAGAU------ -------------.((((((.-.......))))))..(((((((((.....((--((((.....))))))(((.....)))..)))))))))..................------ ( -28.60) >DroSec_CAF1 32308 94 - 1 -------------UAGCUAGU-CAACUACUUAGCUACUGGCCCACUCAAAUCA--CAAUUUAAAGUUGUGGGUCAGUUGCCUUAGUGGGGCAAUAAACUAAUACGCAGAU------ -------------(((((((.-.......)))))))(((((((((........--............)))))))))((((((.....)))))).................------ ( -27.55) >DroSim_CAF1 30217 94 - 1 -------------CAGCUAGU-CAACUACUUAGCUACUGGCCCACUCAAAUCA--CAAUUUAAAGUUGUGGGUCAGUUGCCUUAGUGGGCCAAUAAACUAAUACGCAGAU------ -------------.((((((.-.......))))))..(((((((((.....((--((((.....))))))(((.....)))..)))))))))..................------ ( -28.60) >DroEre_CAF1 36738 94 - 1 -------------CAGCUUGU-CAACUACUUAGCUACUGGCCCACUCAAAUCA--CAAUUUAAUGUUGUGGGUCAGUUGCCUUAGUGGGCCAAUAAACUAAUACGCAGAA------ -------------.((((.((-.....))..))))..(((((((((.....((--((((.....))))))(((.....)))..)))))))))..................------ ( -26.80) >DroYak_CAF1 28942 94 - 1 -------------CAGCUAGU-CAACUGCUUAGCUACUGGCCCACUCAAAUCA--CAAUUUAAUGCUGUGGGUCAGCUGCCUUAGUGGGCCAAUAAACUAAUACGCUGAU------ -------------((((((((-...(..((..((..(((((((((........--............)))))))))..))...))..)........))))....))))..------ ( -26.65) >DroAna_CAF1 36927 114 - 1 GCCUUGUUGUGCCUUGCUGGCCCGACU--UUGGCUACUGGCCCACUCAAAUCACACAAUUUAAUGUUGUGGGUCAGCCGCCUUAGUGGGCCAAUAAACUAAUCCGCAGUCUGCCGC (((..((((.(((.....))).)))).--..)))...(((((((((.......((((((.....))))))((....)).....)))))))))............((.....))... ( -39.90) >consensus _____________CAGCUAGU_CAACUACUUAGCUACUGGCCCACUCAAAUCA__CAAUUUAAAGUUGUGGGUCAGUUGCCUUAGUGGGCCAAUAAACUAAUACGCAGAU______ .........................(((((..((.((((((((((......................)))))))))).))...)))))............................ (-22.81 = -23.37 + 0.56)


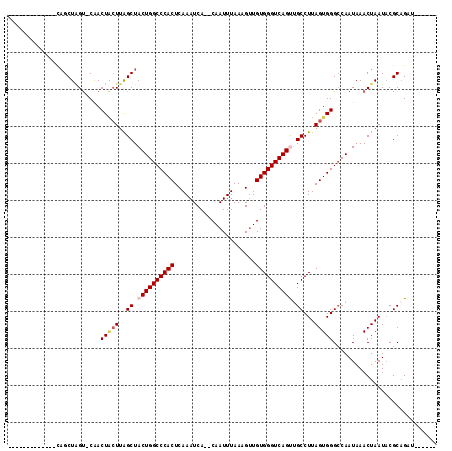
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:50:59 2006