
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,317,091 – 27,317,221 |
| Length | 130 |
| Max. P | 0.998407 |

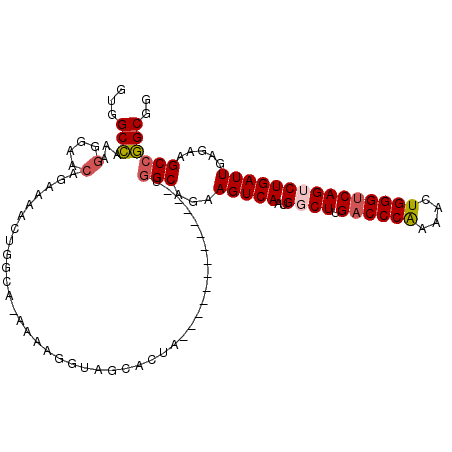
| Location | 27,317,091 – 27,317,188 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.83 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27317091 97 + 27905053 UUGGCCAAAGGGAACAGAAAACUGGCA-AAAAGGUAGCACU--------------UGGCAGAAGUCAAUGGCUUGACCCAUAUCUGGGUCAGUCUGAUUGAGAAGCCGGCGG ...(((.(((....(((....)))((.-........)).))--------------)(((...(((((..((((.((((((....))))))))))))))).....)))))).. ( -29.30) >DroSec_CAF1 21952 98 + 1 GUGGCCAAAGGGAACAGGAAACUGGCA-AAAAGGUAGCACUA-------------UGGCAGAAGUCAAUGUCUUGACCCAAAACUGGGUCAGUCUGAUUGAGCAGCCGGCGG ...(((........(((....)))...-....(((.((....-------------.((((........))))((((((((....)))))))).........)).)))))).. ( -32.80) >DroSim_CAF1 22457 83 + 1 GUGGCCAAAGGGAACA----------------GGUAGCACUA-------------UGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGCAGCCGGCGG ...(((...(....).----------------(((.((....-------------((((....))))..((((.((((((....)))))))))).......)).)))))).. ( -29.70) >DroEre_CAF1 22257 98 + 1 GUGGCCAAAGGGAACAGAAAACUGGGCAG-AAGGUAGCACAA-------------UGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGG ...(((.....(..(((....)))..)..-..(((..(.(((-------------((((....)))...((((.((((((....))))))))))..)))).)..)))))).. ( -31.20) >DroYak_CAF1 18366 99 + 1 GUGGCCAAGGGGAACAGAAAACUGGGCAAAAAGGUAGCACAA-------------UGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGG ...(((.....(..(((....)))..).....(((..(.(((-------------((((....)))...((((.((((((....))))))))))..)))).)..)))))).. ( -31.20) >DroAna_CAF1 24500 105 + 1 GUAGCGAAGUGGA------GAGUGGAG-CCAAGGGAACAGUGCGGAAACCAGCAGAAGCAGAAGUCAAUGGCUUGUCCCGAAACUGGGUCAGACUGAUUGAGGAGCCCGCGG .............------..((((.(-((..((((.(((.(((....)..((....))...........)))))))))....((.((((.....)))).))).)))))).. ( -26.30) >consensus GUGGCCAAAGGGAACAGAAAACUGGCA_AAAAGGUAGCACUA_____________UGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGG ...(((...(....).........................................(((...(((((..((((.((((((....))))))))))))))).....)))))).. (-22.42 = -22.83 + 0.42)

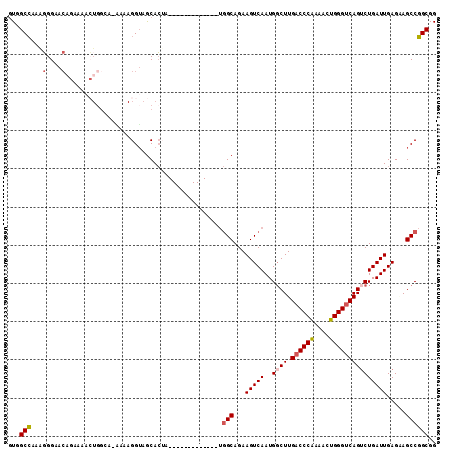
| Location | 27,317,091 – 27,317,188 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27317091 97 - 27905053 CCGCCGGCUUCUCAAUCAGACUGACCCAGAUAUGGGUCAAGCCAUUGACUUCUGCCA--------------AGUGCUACCUUUU-UGCCAGUUUUCUGUUCCCUUUGGCCAA ..((((((((((.....))).(((((((....)))))))))))...((...(((.((--------------((.........))-)).)))...))..........)))... ( -25.20) >DroSec_CAF1 21952 98 - 1 CCGCCGGCUGCUCAAUCAGACUGACCCAGUUUUGGGUCAAGACAUUGACUUCUGCCA-------------UAGUGCUACCUUUU-UGCCAGUUUCCUGUUCCCUUUGGCCAC ..((((((...(((((.....((((((......))))))....))))).....)))(-------------(((.(((..(....-.)..)))...)))).......)))... ( -23.00) >DroSim_CAF1 22457 83 - 1 CCGCCGGCUGCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCA-------------UAGUGCUACC----------------UGUUCCCUUUGGCCAC ..((((((...(((((..(..((((((......))))))..).))))).....)))(-------------(((......)----------------))).......)))... ( -22.90) >DroEre_CAF1 22257 98 - 1 CCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCA-------------UUGUGCUACCUU-CUGCCCAGUUUUCUGUUCCCUUUGGCCAC ..((((((...(((((..(..((((((......))))))..).))))).....)))(-------------(((.((......-..)).))))..............)))... ( -26.20) >DroYak_CAF1 18366 99 - 1 CCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCA-------------UUGUGCUACCUUUUUGCCCAGUUUUCUGUUCCCCUUGGCCAC ..((((((...(((((..(..((((((......))))))..).))))).....)))(-------------(((.((.........)).))))..............)))... ( -25.40) >DroAna_CAF1 24500 105 - 1 CCGCGGGCUCCUCAAUCAGUCUGACCCAGUUUCGGGACAAGCCAUUGACUUCUGCUUCUGCUGGUUUCCGCACUGUUCCCUUGG-CUCCACUC------UCCACUUCGCUAC ..(((((..(((((((..(..((.(((......))).))..).))))).....((....)).))..)))))..........(((-(.......------........)))). ( -22.66) >consensus CCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCA_____________UAGUGCUACCUUUU_GCCCAGUUUUCUGUUCCCUUUGGCCAC ..((((((...(((((.....((((((......))))))....))))).....)))................................(((....)))........)))... (-18.64 = -19.27 + 0.63)

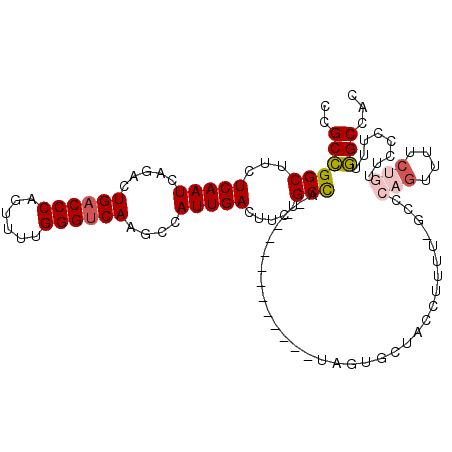

| Location | 27,317,124 – 27,317,221 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -32.52 |
| Energy contribution | -33.20 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27317124 97 + 27905053 UAGCACU--------------UGGCAGAAGUCAAUGGCUUGACCCAUAUCUGGGUCAGUCUGAUUGAGAAGCCGGCGG-------CCGUCGACUGCCAAAAAUGUUAAUUAAUUCGAA (((((.(--------------((((((.(((((..((((.((((((....))))))))))))))).......((((..-------..)))).)))))))...)))))........... ( -33.90) >DroSec_CAF1 21985 98 + 1 UAGCACUA-------------UGGCAGAAGUCAAUGUCUUGACCCAAAACUGGGUCAGUCUGAUUGAGCAGCCGGCGG-------CCGCCGACUGCCAAAAAUGUUAAUUAAUUCGAA (((((...-------------((((((...(((((...((((((((....))))))))....))))).....((((..-------..)))).))))))....)))))........... ( -33.40) >DroSim_CAF1 22475 98 + 1 UAGCACUA-------------UGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGCAGCCGGCGG-------CCGCCGACUGCCAAAAAUGUUAAUUAAUUCGAA (((((...-------------((((((.(((((..((((.((((((....))))))))))))))).......((((..-------..)))).))))))....)))))........... ( -35.90) >DroEre_CAF1 22290 98 + 1 UAGCACAA-------------UGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGG-------CCGUCGACUGCCAAAAAUGUUAAUUAAUUCGAA (((((...-------------((((((.(((((..((((.((((((....))))))))))))))).......((((..-------..)))).))))))....)))))........... ( -33.20) >DroYak_CAF1 18400 105 + 1 UAGCACAA-------------UGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGGCCGGUGGCCGUCGACUGCCAAAAAUGUUAAUUAAUUCGAA (((((...-------------((((((.(((((..((((.((((((....))))))))))))))).......((((((((...)))))))).))))))....)))))........... ( -43.50) >DroAna_CAF1 24527 111 + 1 GAACAGUGCGGAAACCAGCAGAAGCAGAAGUCAAUGGCUUGUCCCGAAACUGGGUCAGACUGAUUGAGGAGCCCGCGG-------CCGUCGACUGCCAAAAAUGUUAAUUAAUUCGAA .((((..((((...((.((....)).....(((((((..((.((((....)))).))..)).)))))))...))))((-------(.(....).))).....))))............ ( -28.60) >consensus UAGCACUA_____________UGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGG_______CCGUCGACUGCCAAAAAUGUUAAUUAAUUCGAA .....................((((((...(((((((.((((((((....)))))))).)).))))).....(((((((.....))))))).)))))).................... (-32.52 = -33.20 + 0.68)



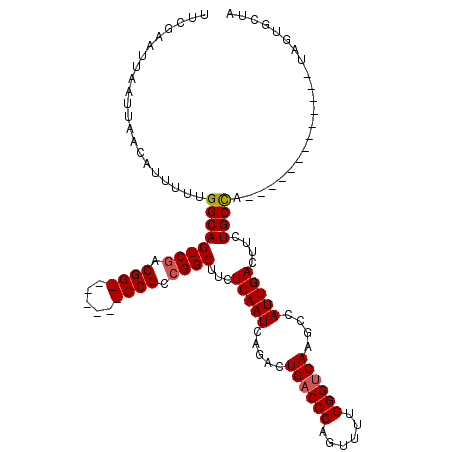
| Location | 27,317,124 – 27,317,221 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -24.60 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27317124 97 - 27905053 UUCGAAUUAAUUAACAUUUUUGGCAGUCGACGG-------CCGCCGGCUUCUCAAUCAGACUGACCCAGAUAUGGGUCAAGCCAUUGACUUCUGCCA--------------AGUGCUA ..................((((((((((((.((-------(........(((.....))).(((((((....))))))).))).)))))...)))))--------------))..... ( -28.80) >DroSec_CAF1 21985 98 - 1 UUCGAAUUAAUUAACAUUUUUGGCAGUCGGCGG-------CCGCCGGCUGCUCAAUCAGACUGACCCAGUUUUGGGUCAAGACAUUGACUUCUGCCA-------------UAGUGCUA ..............((((..((((((..(((((-------((...))))))(((((.....((((((......))))))....))))))..))))))-------------.))))... ( -33.20) >DroSim_CAF1 22475 98 - 1 UUCGAAUUAAUUAACAUUUUUGGCAGUCGGCGG-------CCGCCGGCUGCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCA-------------UAGUGCUA ..............((((..((((((..(((((-------((...))))))(((((..(..((((((......))))))..).))))))..))))))-------------.))))... ( -33.90) >DroEre_CAF1 22290 98 - 1 UUCGAAUUAAUUAACAUUUUUGGCAGUCGACGG-------CCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCA-------------UUGUGCUA ..............(((...((((((((((.((-------(........(((.....))).((((((......)))))).))).)))))...)))))-------------..)))... ( -27.90) >DroYak_CAF1 18400 105 - 1 UUCGAAUUAAUUAACAUUUUUGGCAGUCGACGGCCACCGGCCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCA-------------UUGUGCUA ..............(((...(((((((((.(((((...))))).))))...(((((..(..((((((......))))))..).)))))....)))))-------------..)))... ( -35.60) >DroAna_CAF1 24527 111 - 1 UUCGAAUUAAUUAACAUUUUUGGCAGUCGACGG-------CCGCGGGCUCCUCAAUCAGUCUGACCCAGUUUCGGGACAAGCCAUUGACUUCUGCUUCUGCUGGUUUCCGCACUGUUC ............((((.....(((.(....).)-------))(((((..(((((((..(..((.(((......))).))..).))))).....((....)).))..)))))..)))). ( -26.40) >consensus UUCGAAUUAAUUAACAUUUUUGGCAGUCGACGG_______CCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCA_____________UAGUGCUA .....................((((((((.((((.....)))).))))...(((((.....((((((......))))))....)))))....))))...................... (-24.60 = -25.00 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:50:54 2006