
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,315,283 – 27,315,400 |
| Length | 117 |
| Max. P | 0.931718 |

| Location | 27,315,283 – 27,315,400 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.97 |
| Mean single sequence MFE | -24.19 |
| Consensus MFE | -13.54 |
| Energy contribution | -14.99 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
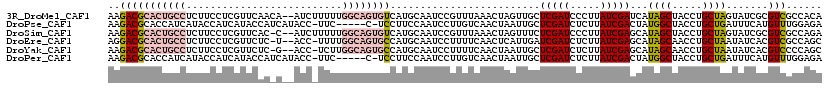
>3R_DroMel_CAF1 27315283 117 - 27905053 AAGACGCACUGCCUCUUCCUCGUUCAACA--AUCUUUUUGGCAGUGUCAUGCAAUCCGUUUAAACUAGUUGCUCGAUCCCUUAUCGAUCAUAGCUACCUGCUAGUAUCGCGUCGCCACA ..(((((((((((................--........))))))(((..(((((..((....))..)))))..)))........(((..((((.....))))..))))))))...... ( -29.86) >DroPse_CAF1 5197 112 - 1 AAGACGCACCAUCAUACCAUCAUACCAUCAUACC-UUC-----C-UCCUUCCAAUCCUUGUCAACUAAUUGCUCGAUCUCUUAUCGACUAUGGCUACCUGCUGAUUUCAUGUUUGGAGA ..................................-...-----.-...((((((.....(((..........(((((.....)))))....(((.....)))))).......)))))). ( -12.00) >DroSim_CAF1 20668 116 - 1 AAGACGCACUGCCUCUUCCUCGUUCAC-C--AUCUUUUUGGCAGUGUCAUGCAAUCCGUUUAAACUAGUUUCUCGAUCCCUUAUCGAGCAUAGCUACCUGCUAGUAUCGCGUCGCCAGA ..(((((((((((..............-.--........))))))...((((...................((((((.....))))))..((((.....)))))))).)))))...... ( -28.80) >DroEre_CAF1 20526 115 - 1 AGGACGCACUGCCUCUUCCUCGUUCUC-U--ACC-UUUUGGCAGUGCCAUGCAAUCCUUUUCAACUCAUUGAUCGAUCUCUUAUCGAGCAUAGCAACCUGCUAAUAUCACGUCGCCAGC ..(((((((((((..............-.--...-....))))))))..(((........((((....))))(((((.....))))))))((((.....)))).......)))...... ( -27.95) >DroYak_CAF1 16624 115 - 1 AAGACGCACUGCCUCUUCCUCGUUCUC-G--ACC-UCUUGGCAGUGCCAUGCAAUCCUUUUCAACUAAUUGCUCGAUCUCUUAUCGAGCAUAGCAACCUGCUAAUAUCACGUCCCCAGC ..(((((((((((......(((....)-)--)..-....))))))))......................((((((((.....))))))))((((.....)))).......)))...... ( -34.50) >DroPer_CAF1 5194 112 - 1 AAGACGCACCAUCAUACCAUCAUACCAUCAUACC-UUC-----C-UCCUUCCAAUCCUUGUCAACUAAUUGCUCGAUCUCUUAUCGACUAUGGCUACCUGCUGAUUUCAUGUUUGGAGA ..................................-...-----.-...((((((.....(((..........(((((.....)))))....(((.....)))))).......)))))). ( -12.00) >consensus AAGACGCACUGCCUCUUCCUCGUUCAA_C__ACC_UUUUGGCAGUGCCAUGCAAUCCUUUUCAACUAAUUGCUCGAUCUCUUAUCGAGCAUAGCUACCUGCUAAUAUCACGUCGCCAGA ..(((((((((((..........................)))))))).........................(((((.....)))))...((((.....)))).......)))...... (-13.54 = -14.99 + 1.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:50:51 2006