| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,208,109 – 3,208,297 |
| Length | 188 |
| Max. P | 0.840892 |

| Location | 3,208,109 – 3,208,218 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 91.51 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -27.06 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
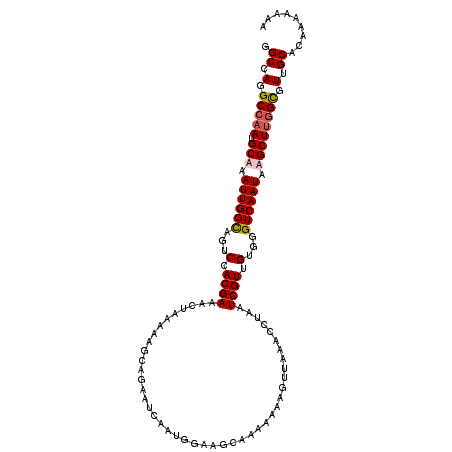
>3R_DroMel_CAF1 3208109 109 - 27905053 GUUUCGUGGACUGCCAAUUUGCAUUGGCCUGGCCAUUAGGAUCGGGUCAGUCAGUAUAAGUUGGAGUUCUUUAUUGUUGUGUGAUAGUCUGAUAGUCGGACCACAGGCA .......(((((.((((((((.((((((.(((((..........))))))))))).))))))))))))).....((((.((((...(((((.....))))))))))))) ( -35.50) >DroSec_CAF1 27499 107 - 1 GUUUCGUGGACUACCAAUUUGCAUUGGCCUGGCCAUUAGGAUCGGGUCAGUCAGU--UAGUUGGAGUUCUUUAUUGUUGUGUGAUAGUCUGAUAGUCGGGACACAGGCA .....(((((((.((((((.((((((((((((((....)).))))))))))..))--.))))))))))).......((((((.....((((.....)))))))))))). ( -33.70) >DroSim_CAF1 34318 107 - 1 GUUUCGUUGACUGCCAAUUUGCAUUGGCCUGGCCAUUAGGAUCGGGUCAGUCAGU--AAGUUGGAGUUCUUUAUUGUUGUGUGAUAGUCUGAUAGUCGGGACACAGGCA ((.((.((((((((((((((((((((((((((((....)).))))))))))..))--)))))))...((...((((((....))))))..))))))))))).))..... ( -34.40) >DroEre_CAF1 27000 99 - 1 GUUUCGUGGACUGCCAAUUUGCAUUGGCCUGGCCAUUAGCAUCGGGUCAGUCAGU--AAGUUGGAGUUCUUUAUUGUUGUGUGAU--------AGUUGGGCCACAGACA ((((.(((((((.((((((((((((((((((((.....))..)))))))))..))--)))))))))).((..((((((....)))--------)))..)))))))))). ( -34.10) >consensus GUUUCGUGGACUGCCAAUUUGCAUUGGCCUGGCCAUUAGGAUCGGGUCAGUCAGU__AAGUUGGAGUUCUUUAUUGUUGUGUGAUAGUCUGAUAGUCGGGACACAGGCA .....(((((((.(((((((((((((((((((((....)).))))))))))..))..))))))))))))....((((.((.((((.........)))).)).)))))). (-27.06 = -27.75 + 0.69)

| Location | 3,208,149 – 3,208,257 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -21.67 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3208149 108 - 27905053 AGGUUCAACUUUUUUUGCUUUCAUUGAUUCUGCUUUUUAGUUUCGUGGACUGCCAAUUUGCAUUGGCCUGGCCAUUAGGAUCGGGUCAGUCAGUAUAAGUUGGAGUUC ((.((((((((....((((..(((.((..(((.....)))..)))))((((((......))).((((((((((....)).))))))))))))))).)))))))).)). ( -28.80) >DroSec_CAF1 27539 106 - 1 AGGUUUAUCUAUUUUUGCUUCCAUUGAUUCUGCUUUUUAGUUUCGUGGACUACCAAUUUGCAUUGGCCUGGCCAUUAGGAUCGGGUCAGUCAGU--UAGUUGGAGUUC ...................(((((.((..(((.....)))..)))))))((.((((((.((((((((((((((....)).))))))))))..))--.))))))))... ( -29.00) >DroSim_CAF1 34358 106 - 1 AGGUUUAACUUUUUUUGCUUCCAUUGAUUCUGCUUUUUAGUUUCGUUGACUGCCAAUUUGCAUUGGCCUGGCCAUUAGGAUCGGGUCAGUCAGU--AAGUUGGAGUUC .....................((.(((..(((.....)))..))).))(((.(((((((((((((((((((((....)).))))))))))..))--)))))))))).. ( -29.90) >DroEre_CAF1 27032 89 - 1 AGGUUUAACUUUUU-UGCU----------------UUUAGUUUCGUGGACUGCCAAUUUGCAUUGGCCUGGCCAUUAGCAUCGGGUCAGUCAGU--AAGUUGGAGUUC ..............-....----------------...........(((((.((((((((((((((((((((.....))..)))))))))..))--)))))))))))) ( -26.00) >consensus AGGUUUAACUUUUUUUGCUUCCAUUGAUUCUGCUUUUUAGUUUCGUGGACUGCCAAUUUGCAUUGGCCUGGCCAUUAGGAUCGGGUCAGUCAGU__AAGUUGGAGUUC ..............................................(((((.(((((((((((((((((((((....)).))))))))))..))..)))))))))))) (-21.67 = -22.43 + 0.75)


| Location | 3,208,184 – 3,208,297 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.62 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3208184 113 + 27905053 GGCCAGGCCAAUGCAAAUUGGCAGUCCACGAAACUAAAAAGCAGAAUCAAUGAAAGCAAAAAAAGUUGAACCUAAUCGUUGUGGGUCAAUAAGCUUAGCGUUGCACAAAAAAA ((....((((((....))))))...))....................(((((.((((.......(((((.((((.......)))))))))..))))..))))).......... ( -20.70) >DroSec_CAF1 27572 113 + 1 GGCCAGGCCAAUGCAAAUUGGUAGUCCACGAAACUAAAAAGCAGAAUCAAUGGAAGCAAAAAUAGAUAAACCUAAUCGUUGUGGGUCAAUAUGCUUGGCGUUGCACAAAAAAA .((.(.(((((.(((.(((((...(((((((.........((.............)).....(((......)))....)))))))))))).)))))))).).))......... ( -26.02) >DroSim_CAF1 34391 113 + 1 GGCCAGGCCAAUGCAAAUUGGCAGUCAACGAAACUAAAAAGCAGAAUCAAUGGAAGCAAAAAAAGUUAAACCUAAUCGUUGUGGGUCAAUAUGCUUGGCGUUGCACAAAAAAA .((.(.(((((.(((.((((((...((((((.........((.............))......((......))..))))))...)))))).)))))))).).))......... ( -28.92) >DroEre_CAF1 27065 94 + 1 GGCCAGGCCAAUGCAAAUUGGCAGUCCACGAAACUAAA----------------AGCA-AAAAAGUUAAACCUAAUCGUUGUGGGUCAAUAAGCU-GGUAUUGCACAAA-AAA .(((((((((((....)))))).((((((((.......----------------(((.-.....)))...........))))))).)......))-)))..........-... ( -21.97) >consensus GGCCAGGCCAAUGCAAAUUGGCAGUCCACGAAACUAAAAAGCAGAAUCAAUGGAAGCAAAAAAAGUUAAACCUAAUCGUUGUGGGUCAAUAAGCUUGGCGUUGCACAAAAAAA .((.(.(((((.(((.((((((...(.((((............................................)))).)...)))))).)))))))).).))......... (-17.00 = -17.62 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:27 2006