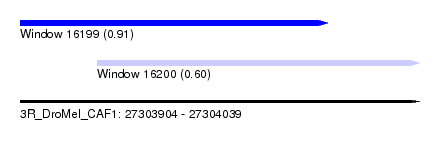
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,303,904 – 27,304,039 |
| Length | 135 |
| Max. P | 0.910383 |

| Location | 27,303,904 – 27,304,008 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -35.69 |
| Consensus MFE | -23.59 |
| Energy contribution | -23.46 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
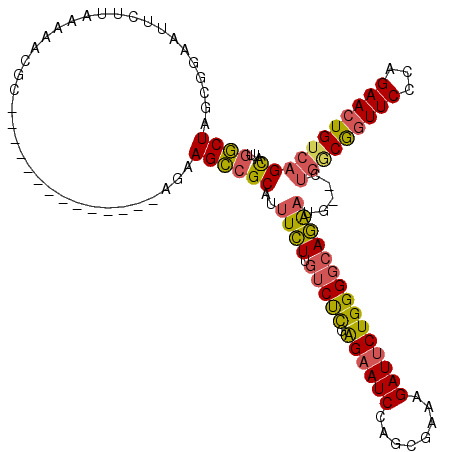
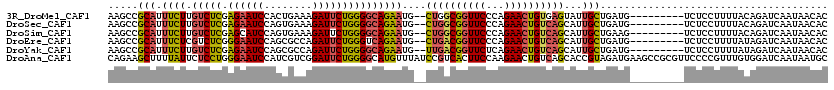
>3R_DroMel_CAF1 27303904 104 + 27905053 GGCUAGCGGAAUUCUUAAAAAUGC--------------AGAAGCCGCAUUUCUUGUCUCGAGAAUCCACUGAAAGAUUCUGGGGCAGAAUG--CUGGCGGUUCCCAGAACUGUGAGUAUU .....((((..((((.........--------------)))).)))).....(((((((.((((((........)))))))))))))((((--((.(((((((...))))))).)))))) ( -34.60) >DroSec_CAF1 8874 104 + 1 GGCUAGCGGAAUUCCUAAAAACGC--------------AGAAGCCGCAUUUCUUGUCUCGAGAAUCCAGUGAAAGAUUCUGGGGCAGAAUG--CUGGCGGUUCCCAGAACUGUCAGCAUU ((((.(((.............)))--------------...)))).......(((((((.((((((........)))))))))))))((((--((((((((((...)))))))))))))) ( -40.72) >DroSim_CAF1 9297 104 + 1 GGCUAGCGGAAUUCUUAAAAACGC--------------AGAAGCCGCAUUUCUUGUCUCGAGCAUCCAGUGAAAGAUUCUGGGGCAGAAUG--CUGGCGGUUCCCAGAACUGUCAGCAUU ((((.(((.............)))--------------...)))).......(((((((.((.(((........))).)))))))))((((--((((((((((...)))))))))))))) ( -36.32) >DroEre_CAF1 9114 104 + 1 GGCUAUCGGAAUUCUUUAAAAUGC--------------AGAAGCCGCAUUUCUCGUCUCGGGAAUCCAGCGCCAGAUUCUGGGUCAGAAUG--CUGACGGUUCCCAGAACUGUCAGCAUU ((((..(((((((.........((--------------....))(((..((((((...))))))....)))...)))))))))))..((((--((((((((((...)))))))))))))) ( -35.70) >DroYak_CAF1 5174 104 + 1 GGCUAGCGGAAUUCUUUAAAAAGC--------------AGAAGCCGCAUUUCUUGUCUCGAGAAUCCAGCGCCAGAUUCUGGGGCAGAAUG--UUGACGGUUCUCAGAACUGUCAGCAUU .....((((..((((.........--------------)))).)))).....(((((((.((((((........)))))))))))))((((--((((((((((...)))))))))))))) ( -38.00) >DroAna_CAF1 10279 120 + 1 GUCUAGACGAGUUCCCUAAAACGCCAAGACAGUUAUGCGGCAGAAGCUUUUAUUCUCCUGGGAAUCCAUCGUCGGAUUCUGGGGCAUGUUUAUCCGUCACUUCCAAGAACUGUCAGCACC ......................((...(((((((....((.((..((...((..(.(((.(((((((......))))))).)))...)..))...))..)).))...))))))).))... ( -28.80) >consensus GGCUAGCGGAAUUCUUAAAAACGC______________AGAAGCCGCAUUUCUUGUCUCGAGAAUCCAGCGAAAGAUUCUGGGGCAGAAUG__CUGGCGGUUCCCAGAACUGUCAGCAUU ((((.....................................))))((..((((.(((((.((((((........))))))))))))))).....(((((((((...)))))))))))... (-23.59 = -23.46 + -0.13)

| Location | 27,303,930 – 27,304,039 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -22.80 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27303930 109 + 27905053 AAGCCGCAUUUCUUGUCUCGAGAAUCCACUGAAAGAUUCUGGGGCAGAAUG--CUGGCGGUUCCCAGAACUGUGAGUAUUGCUGAUG---------UCUCCUUUUACAGAUCAAUAACAC ..(((((((((..((((((.((((((........)))))))))))))))))--).)))........((.(((((((....((....)---------).....))))))).))........ ( -32.30) >DroSec_CAF1 8900 109 + 1 AAGCCGCAUUUCUUGUCUCGAGAAUCCAGUGAAAGAUUCUGGGGCAGAAUG--CUGGCGGUUCCCAGAACUGUCAGCAUUGCUGAUG---------UCUCCUUUUACAGAUCAAUAACAC .(((........(((((((.((((((........)))))))))))))((((--((((((((((...)))))))))))))))))...(---------(((........))))......... ( -35.30) >DroSim_CAF1 9323 109 + 1 AAGCCGCAUUUCUUGUCUCGAGCAUCCAGUGAAAGAUUCUGGGGCAGAAUG--CUGGCGGUUCCCAGAACUGUCAGCAUUGCUGAAG---------UCUCCUUUUACAGAUCAAUAACAC ..((((((.....))...)).)).....(((((((...((..((((..(((--((((((((((...)))))))))))))))))..))---------....)))))))............. ( -32.80) >DroEre_CAF1 9140 109 + 1 AAGCCGCAUUUCUCGUCUCGGGAAUCCAGCGCCAGAUUCUGGGUCAGAAUG--CUGACGGUUCCCAGAACUGUCAGCAUUGCUGAUG---------UCUCCUUUUAUAGAUCAAUAACAC ....(((..((((((...))))))....)))((((...))))(((((((((--((((((((((...)))))))))))))).)))))(---------(((........))))......... ( -37.30) >DroYak_CAF1 5200 109 + 1 AAGCCGCAUUUCUUGUCUCGAGAAUCCAGCGCCAGAUUCUGGGGCAGAAUG--UUGACGGUUCUCAGAACUGUCAGCAUUGCUGAUG---------UCUCCUUUUAUAGAUCAAUAACAC .(((........(((((((.((((((........)))))))))))))((((--((((((((((...)))))))))))))))))...(---------(((........))))......... ( -33.40) >DroAna_CAF1 10319 120 + 1 CAGAAGCUUUUAUUCUCCUGGGAAUCCAUCGUCGGAUUCUGGGGCAUGUUUAUCCGUCACUUCCAAGAACUGUCAGCACCGUAGAUGAAGCCGCGUUCCCCGUUUGUGGAUCAAUAAUGC ((.((((.............(((((((......)))))))((((.(((((((((((...((..((.....))..))...))..)))))....)))).)))))))).))............ ( -26.40) >consensus AAGCCGCAUUUCUUGUCUCGAGAAUCCAGCGAAAGAUUCUGGGGCAGAAUG__CUGGCGGUUCCCAGAACUGUCAGCAUUGCUGAUG_________UCUCCUUUUACAGAUCAAUAACAC .....(((.((((.(((((.((((((........)))))))))))))))....((((((((((...))))))))))...)))...................................... (-22.80 = -22.80 + 0.00)
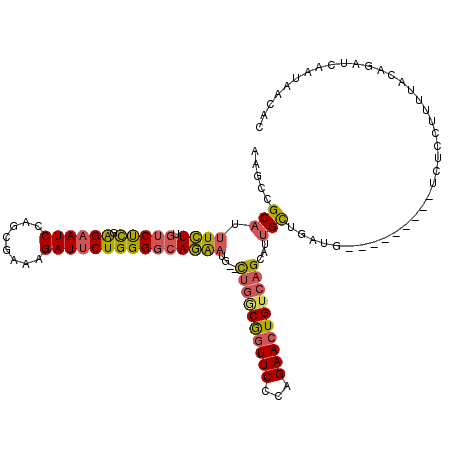
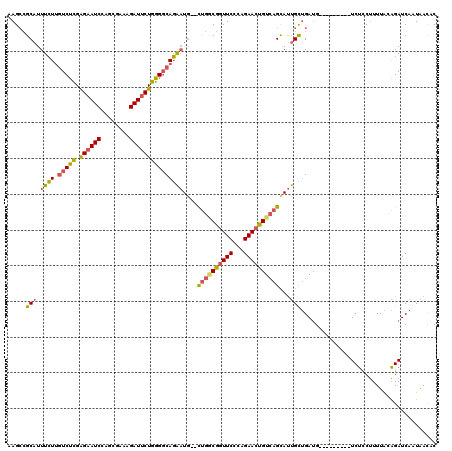
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:50:38 2006