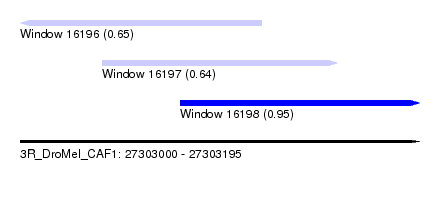
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,303,000 – 27,303,195 |
| Length | 195 |
| Max. P | 0.951682 |

| Location | 27,303,000 – 27,303,118 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.19 |
| Mean single sequence MFE | -37.95 |
| Consensus MFE | -29.76 |
| Energy contribution | -30.02 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27303000 118 - 27905053 AAUUGAGUGAUAACAGCUUUCGCUGUUGGCGGUUGUUAACAUCUAUAAAAUGGAAAGUGUCACAC-CAACGUGACGCCC-AAACAGCCUUGAAAGUGGUUGUUGUUUUUGAGCUGUCGGG ...........((((((....))))))(((((((...(((..........(((...(((((((..-....)))))))))-)(((((((........))))))))))....)))))))... ( -36.20) >DroSec_CAF1 7975 118 - 1 AAUGGAGUGAUAACAGCUUUCGCUGUUGACAGCUGUUAGCGUCUAUAAAAUGGAAAGUGUCACAC-CAACGUGACGCCC-AAACAGCCUUGAAAGUGGUUGUUGUCUUCCAGCUGUCGGG ...........((((((....))))))((((((((..(((..........(((...(((((((..-....)))))))))-)(((((((........)))))))).))..))))))))... ( -44.00) >DroSim_CAF1 8392 118 - 1 AAUGGAGUGAUAACAGCUUUCGCUGUUGACAGCUGUUAGCGUCUAUAAAAUGGAAAGUGUCACAC-CAACGUGACGCCC-AAACAGCCUUGAAAGUGGUUGUUGUCUUCCAGCUGUCGGG ...........((((((....))))))((((((((..(((..........(((...(((((((..-....)))))))))-)(((((((........)))))))).))..))))))))... ( -44.00) >DroEre_CAF1 8244 118 - 1 AAUGGAGUGAUAACAGCUUUCGCUGUUGACGGCUGUUAAUGUCCAUAAAAUGGAAAGUGUCACAC-CAACGUGACGCCC-AAACAACCUUGAAAGUGGUUGUUGUCUUCGAGCUGGCGGG .(((((....((((((((.(((....))).))))))))...)))))..........(((((((..-....)))))))((-.(((((((........)))))))(((........))))). ( -38.90) >DroYak_CAF1 4293 118 - 1 AAUGGAGUGAUAACAGCUUUCGCUGUUGUCGGCUGUAAACGUUCAUAAAAUGGAAAGUGUCACAC-CAACGUGACGCCC-AAACAACCUUGAAAGUGGUUGUUGUCUUCCAGCUUUUGGG ..((.((((((((((((....))))))))).))).)).....(((.((..(((((.(((((((..-....)))))))..-.(((((((........)))))))...)))))..)).))). ( -40.50) >DroAna_CAF1 9311 113 - 1 AAUUGAGCGAUAACUGCUUUCGCUAUUAACACCC------AUCUA-AAAAUGGAAAGUGUCACACACGACAUGACGCCCGAAACAGCCUUGAAACUGGUUUGUGUCAUCAAGUUGUUGGC ....(((((.....)))))..((((.((((..((------((...-...))))...(((((......)))))(((((..(((.(((........))).)))))))).....)))).)))) ( -24.10) >consensus AAUGGAGUGAUAACAGCUUUCGCUGUUGACAGCUGUUAACGUCUAUAAAAUGGAAAGUGUCACAC_CAACGUGACGCCC_AAACAGCCUUGAAAGUGGUUGUUGUCUUCCAGCUGUCGGG ...........((((((....))))))(((((((.......((((.....))))..(((((((.......)))))))....(((((((........))))))).......)))))))... (-29.76 = -30.02 + 0.25)


| Location | 27,303,040 – 27,303,155 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -34.78 |
| Consensus MFE | -24.91 |
| Energy contribution | -25.38 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27303040 115 + 27905053 -GGGCGUCACGUUG-GUGUGACACUUUCCAUUUUAUAGAUGUUAACAACCGCCAACAGCGAAAGCUGUUAUCACUCAAUUCCCAUA---UACCUUGUUCUUUGUUGUGGGACACCCACCU -.(((((((((...-.))))))..((..((((.....))))..)).....)))((((((....))))))..........(((((((---...............)))))))......... ( -32.06) >DroSec_CAF1 8015 115 + 1 -GGGCGUCACGUUG-GUGUGACACUUUCCAUUUUAUAGACGCUAACAGCUGUCAACAGCGAAAGCUGUUAUCACUCCAUUCCCUUA---UACCUGGUUUUUUGUUGUGGGACACCCACCU -(((.(((....((-(.((((................(((((.....)).)))((((((....)))))).)))).)))...((...---.....)).............))).))).... ( -34.80) >DroSim_CAF1 8432 115 + 1 -GGGCGUCACGUUG-GUGUGACACUUUCCAUUUUAUAGACGCUAACAGCUGUCAACAGCGAAAGCUGUUAUCACUCCAUUCCCUUA---UACCUGGUUCUUUGUUGUGGGACACCCACCU -(((.(((....((-(.((((................(((((.....)).)))((((((....)))))).)))).)))...((...---.....)).............))).))).... ( -34.80) >DroEre_CAF1 8284 115 + 1 -GGGCGUCACGUUG-GUGUGACACUUUCCAUUUUAUGGACAUUAACAGCCGUCAACAGCGAAAGCUGUUAUCACUCCAUUCCCAAA---UGUCUGGUUCUUUGUUGUGGGACACCCACUG -.(((((((((...-.))))))....(((((...)))))........)))(((((((((....)))))).((((..((...(((..---....))).....))..)))))))........ ( -35.80) >DroYak_CAF1 4333 115 + 1 -GGGCGUCACGUUG-GUGUGACACUUUCCAUUUUAUGAACGUUUACAGCCGACAACAGCGAAAGCUGUUAUCACUCCAUUCCCAAA---UAUCGUGUUCUUUGUUUUGGGACACCCACCU -(((.(((..((((-((((((.((.(((........))).))))))).)))))((((((....))))))............(((((---...((.......)).)))))))).))).... ( -36.10) >DroAna_CAF1 9351 112 + 1 CGGGCGUCAUGUCGUGUGUGACACUUUCCAUUUU-UAGAU------GGGUGUUAAUAGCGAAAGCAGUUAUCGCUCAAUUCUGAACCAUCAUCGGGGGCUUUGUUG-GGAACACCCACCG ((((((((((.......))))).....((...((-(((((------(((((.((((.((....)).)))).))))))..))))))((......)))))).....((-((....))))))) ( -35.10) >consensus _GGGCGUCACGUUG_GUGUGACACUUUCCAUUUUAUAGACGUUAACAGCCGUCAACAGCGAAAGCUGUUAUCACUCCAUUCCCAAA___UACCUGGUUCUUUGUUGUGGGACACCCACCU .(((.((((((.....))))))...((((((......((((........))))((((((....))))))....................................))))))..))).... (-24.91 = -25.38 + 0.48)



| Location | 27,303,078 – 27,303,195 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.33 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -14.34 |
| Energy contribution | -15.57 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27303078 117 + 27905053 GUUAACAACCGCCAACAGCGAAAGCUGUUAUCACUCAAUUCCCAUA---UACCUUGUUCUUUGUUGUGGGACACCCACCUGUGGAUGAUCACCAGCUACUAGAUGAUUAUCCUUUCACUU ....(((......((((((....))))))..........(((((((---...............)))))))........)))(((((((((.(........).)))))))))........ ( -29.96) >DroSec_CAF1 8053 117 + 1 GCUAACAGCUGUCAACAGCGAAAGCUGUUAUCACUCCAUUCCCUUA---UACCUGGUUUUUUGUUGUGGGACACCCACCUGCGGAUGAUCACCAGCUACUGGAUGAUGAUCCUUUCACUU ((.......((((((((((....)))))).((((..((...((...---.....)).....))..)))))))).......))((((.((((((((...)))).)))).))))........ ( -36.14) >DroSim_CAF1 8470 117 + 1 GCUAACAGCUGUCAACAGCGAAAGCUGUUAUCACUCCAUUCCCUUA---UACCUGGUUCUUUGUUGUGGGACACCCACCUGUGGAUGAUCACCAGCUACUGGAUGAUGAUCCUUUCACUU ....((((.((((((((((....)))))).((((..((...((...---.....)).....))..)))))))).....))))((((.((((((((...)))).)))).))))........ ( -35.80) >DroEre_CAF1 8322 100 + 1 AUUAACAGCCGUCAACAGCGAAAGCUGUUAUCACUCCAUUCCCAAA---UGUCUGGUUCUUUGUUGUGGGACACCCACUG--------------GCUACUGGAUG---AUCCUUUCACUU ......(((((..((((((....))))))..).........(((..---....))).........(((((...))))).)--------------)))...((...---..))........ ( -26.00) >DroYak_CAF1 4371 100 + 1 GUUUACAGCCGACAACAGCGAAAGCUGUUAUCACUCCAUUCCCAAA---UAUCGUGUUCUUUGUUUUGGGACACCCACCU--------------GCUACUGGAUG---AUCCUUUCACUU ......(((.((.((((((....)))))).)).......(((((((---...((.......)).))))))).........--------------)))...((...---..))........ ( -23.90) >DroAna_CAF1 9390 111 + 1 ------GGGUGUUAAUAGCGAAAGCAGUUAUCGCUCAAUUCUGAACCAUCAUCGGGGGCUUUGUUG-GGAACACCCACCGAAAGAUGAUCGGUGGCUACUGA--GAAGAUCCUUUCACUU ------(((((.((((.((....)).)))).))))).....((((((......))((..(((.(..-(......(((((((.......)))))))...)..)--.)))..)).))))... ( -34.00) >consensus GUUAACAGCCGUCAACAGCGAAAGCUGUUAUCACUCCAUUCCCAAA___UACCUGGUUCUUUGUUGUGGGACACCCACCUG_GGAUGAUCACCAGCUACUGGAUGAUGAUCCUUUCACUU ......(((....((((((....))))))....................................((((.....))))................)))...((((....))))........ (-14.34 = -15.57 + 1.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:50:36 2006