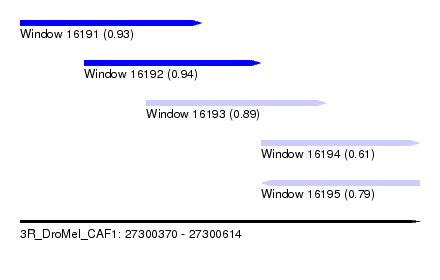
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,300,370 – 27,300,614 |
| Length | 244 |
| Max. P | 0.938719 |

| Location | 27,300,370 – 27,300,481 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -25.21 |
| Energy contribution | -25.77 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27300370 111 + 27905053 ACACGACUUUCAUGUUAGCAAGUCGUUC-GCUAACAGUAA-AAUCAAUAAGUACCAGGCGGCUUAUUAUGAGCCUUGCU-------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAU ........(((.((((((((((..((((-((.....))..-....(((((((........)))))))..))))))))))-------))))(((((((((.....))))))))).)))... ( -34.70) >DroSec_CAF1 5377 111 + 1 ACACGACUUUCAUGUUAGCAAGUCGUUC-GCUAACAGUAA-AAUCAAUAAGUACCAGGCGGCUUAUUAUGAGCCUUGCU-------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAU ........(((.((((((((((..((((-((.....))..-....(((((((........)))))))..))))))))))-------))))(((((((((.....))))))))).)))... ( -34.70) >DroSim_CAF1 5798 111 + 1 ACACGACUUUCAUGUUAGCAAGUCGUUC-GCUAACAGUAA-AAUCAAUAAGUACCAGGCGGCUUAUUAUGAGCCUUGCU-------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAU ........(((.((((((((((..((((-((.....))..-....(((((((........)))))))..))))))))))-------))))(((((((((.....))))))))).)))... ( -34.70) >DroEre_CAF1 5517 112 + 1 ACACGACUUUCAUGUUAGCAAGUCGUUC-UCUAACAGUAAAAAUCAAUAUGUACCAGGCGGCUUAUUAUGAGCCUUGCC-------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAU ....(((((.(......).)))))((((-(..........................((((((((.....)))))..)))-------....(((((((((.....)))))))))))))).. ( -33.30) >DroYak_CAF1 1509 119 + 1 ACACGACUUUCAUGUUAGCAAGUCGUUC-GCUAACAGUAAAAAUCAAUAAGUACCAGGCGGCUUAUUAUGAGCCUUGCUAACUGCUAACAGUGAGCUAAAUAACUUGGCUCACAGAACAU ........(((..(((((((((..((((-((.....)).......(((((((........)))))))..)))))))))))))........(((((((((.....))))))))).)))... ( -33.20) >DroAna_CAF1 6050 112 + 1 ACACGACUUUCAUGUUAGCAAGUCGUUGUUGUAACAGUAAAAAUCAAUAAGUACCAGGCUGCCUAUUAUGAGCC-AGCG-------AACAGUGAACUAAAUAACUUGGCUCACAGAACAU ..(((((((.(......).)))))))((((.(..((((......(.....)......)))).......((((((-((..-------...((....)).......)))))))).).)))). ( -21.90) >consensus ACACGACUUUCAUGUUAGCAAGUCGUUC_GCUAACAGUAA_AAUCAAUAAGUACCAGGCGGCUUAUUAUGAGCCUUGCU_______AACAGUGAGCUAAAUAACUUGGCUCACAGAACAU ..(((((((.(......).))))))).................((...........((((((((.....)))))..)))...........(((((((((.....))))))))).)).... (-25.21 = -25.77 + 0.56)
| Location | 27,300,409 – 27,300,517 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.53 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -25.64 |
| Energy contribution | -26.78 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27300409 108 + 27905053 -AAUCAAUAAGUACCAGGCGGCUUAUUAUGAGCCUUGCU-------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGGCAAUCAUCGUCGUU---CGCCUUUUGUACC-CCCUC -.........((((.(((((((((.....))))).....-------....(((((((((.....))))))))).((((..((((......)))).)))---)))))...)))).-..... ( -34.10) >DroSec_CAF1 5416 105 + 1 -AAUCAAUAAGUACCAGGCGGCUUAUUAUGAGCCUUGCU-------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGGCAAUCAUCGUCGUU---CGCCUUUUGUACC-CC--- -.........((((.(((((((((.....))))).....-------....(((((((((.....))))))))).((((..((((......)))).)))---)))))...)))).-..--- ( -34.10) >DroSim_CAF1 5837 105 + 1 -AAUCAAUAAGUACCAGGCGGCUUAUUAUGAGCCUUGCU-------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGGCAAUCAUCGUCGUU---CGCCUUUUGUACC-CC--- -.........((((.(((((((((.....))))).....-------....(((((((((.....))))))))).((((..((((......)))).)))---)))))...)))).-..--- ( -34.10) >DroEre_CAF1 5556 106 + 1 AAAUCAAUAUGUACCAGGCGGCUUAUUAUGAGCCUUGCC-------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGGCAAUCAUCUUCGUU---CGCCUUUUGUACC-CC--- ..........((((.(((((((((.....)))))(((((-------....(((((((((.....))))))))).(......))))))...........---.))))...)))).-..--- ( -33.90) >DroYak_CAF1 1548 114 + 1 AAAUCAAUAAGUACCAGGCGGCUUAUUAUGAGCCUUGCUAACUGCUAACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGCCAAUCAUCGUCGCU---CGCCUUCUGUUCCCCC--- ...............(((((((((.....)))))..((.....)).....(((((((((.....))))))))).......(((.(.......).))).---.))))...........--- ( -28.00) >DroAna_CAF1 6090 103 + 1 AAAUCAAUAAGUACCAGGCUGCCUAUUAUGAGCC-AGCG-------AACAGUGAACUAAAUAACUUGGCUCACAGAACAUGCAGCAAUCAUCGUUGAUGCUCGUCU-----CCC-CC--- ..(((((..(.......(((((......((((((-((..-------...((....)).......))))))))........))))).....)..)))))........-----...-..--- ( -22.14) >consensus _AAUCAAUAAGUACCAGGCGGCUUAUUAUGAGCCUUGCU_______AACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGGCAAUCAUCGUCGUU___CGCCUUUUGUACC_CC___ ..........((((.(((((((((.....)))))................(((((((((.....))))))))).......(((((.......))))).....))))...))))....... (-25.64 = -26.78 + 1.14)
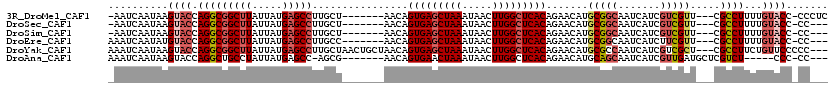

| Location | 27,300,447 – 27,300,557 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.65 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27300447 110 + 27905053 ------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGGCAAUCAUCGUCGUU---CGCCUUUUGUACC-CCCUCCCUCUGCUCUACAAUAGUCCAUCCAAUAGGCAUUAAUUUU ------....(((((((((.....))))))))).((((..((((......)))).)))---)((((.(((....-............................))).))))......... ( -24.05) >DroSec_CAF1 5454 106 + 1 ------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGGCAAUCAUCGUCGUU---CGCCUUUUGUACC-CC----CCCUGCUCUACAAUAGGCCAUCCAAUAGUCAUUAAUUUU ------....(((((((((.....))))))))).((((..((((......)))).)))---)((((.(((((..-..----........))))).))))..................... ( -26.90) >DroSim_CAF1 5875 106 + 1 ------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGGCAAUCAUCGUCGUU---CGCCUUUUGUACC-CC----CUCUGCUCUACAAUAGGCCAUCCAAUAGGCAUUAAUUUU ------....(((((((((.....))))))))).((((..((((......)))).)))---)((((.(((....-.(----((.(........).))).....))).))))......... ( -27.80) >DroEre_CAF1 5595 96 + 1 ------AACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGGCAAUCAUCUUCGUU---CGCCUUUUGUACC-CC----CUCUG----------AGCAGUCGAAUAGGCAUUAAUUUU ------....(((((((((.....))))))))).((((.................)))---)((((((((.((.-.(----.....----------.)..)))))).))))......... ( -21.73) >DroYak_CAF1 1588 103 + 1 ACUGCUAACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGCCAAUCAUCGUCGCU---CGCCUUCUGUUCCCCC----CACUG----------AACAGUCCAAUAGGCAUUAAUUUU ..........(((((((((.....))))))))).......(((.(.......).))).---.((((.((((((....----....)----------)))))......))))......... ( -26.90) >DroAna_CAF1 6128 93 + 1 ------AACAGUGAACUAAAUAACUUGGCUCACAGAACAUGCAGCAAUCAUCGUUGAUGCUCGUCU-----CCC-CC----UUCU-----------AACAAGCCAUUAGGCAUUAAUUUU ------....((((.((((.....)))).)))).....(((.((((.(((....))))))))))..-----...-..----....-----------.....(((....)))......... ( -17.10) >consensus ______AACAGUGAGCUAAAUAACUUGGCUCACAGAACAUGCGGCAAUCAUCGUCGUU___CGCCUUUUGUACC_CC____CUCUG__________AGCAAUCCAAUAGGCAUUAAUUUU ..........(((((((((.....))))))))).......(((((.......))))).....((((.........................................))))......... (-16.31 = -16.65 + 0.34)
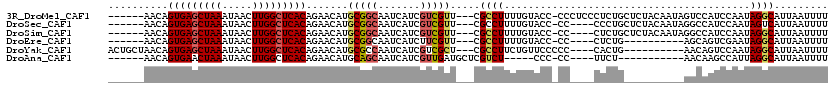
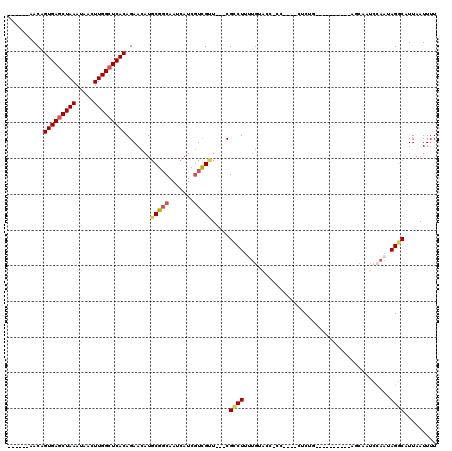
| Location | 27,300,517 – 27,300,614 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -16.75 |
| Consensus MFE | -8.98 |
| Energy contribution | -9.60 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27300517 97 + 27905053 CCUCUGCUCUACAAUAGUCCAUCCAAUAGGCAUUAAUUUUAGCCAUAUGCACUGGACUAUUCCAUACACUAUACACCA--------------UAUAGCAUACACCAGGGA----C .(((((......(((((((((....((((((..........))).)))....))))))))).......(((((.....--------------))))).......))))).----. ( -21.60) >DroSec_CAF1 5521 110 + 1 -CCCUGCUCUACAAUAGGCCAUCCAAUAGUCAUUAAUUUUAGCCAUAUGCACUGGACUAUUCCAUACACCAUACACCAUACACCAUAUAGCAUAUAGCAUACACCAGGGA----C -(((((..........(((.........)))..........((.((((((..((((....)))).............(((.....))).)))))).))......))))).----. ( -19.00) >DroSim_CAF1 5942 96 + 1 -CUCUGCUCUACAAUAGGCCAUCCAAUAGGCAUUAAUUUUAGCCAUAUGCACUGGACUAUUCCAUACACCAUACACCA--------------UAUAGCAUACACCAGGGA----C -(((((...........(((........)))..........((.(((((...((((....))))............))--------------))).))......))))).----. ( -14.86) >DroYak_CAF1 1662 97 + 1 -CACUG----------AACAGUCCAAUAGGCAUUAAUUUUAGCCAUAUGCACUAGACUAUUCCAUACACCAUACACCAUACAAC-------AUAUAGCAUACACCAGGGACCGAC -.....----------..(.((((....(((..........))).(((((..................................-------.....)))))......)))).).. ( -11.53) >consensus _CUCUGCUCUACAAUAGGCCAUCCAAUAGGCAUUAAUUUUAGCCAUAUGCACUGGACUAUUCCAUACACCAUACACCA______________UAUAGCAUACACCAGGGA____C .(((((......................(((..........))).(((((..((((....))))((.....)).......................)))))...)))))...... ( -8.98 = -9.60 + 0.62)

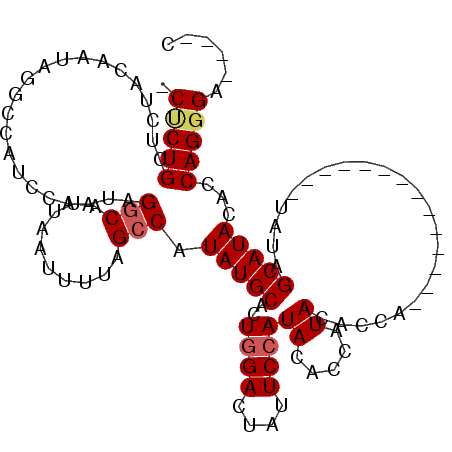
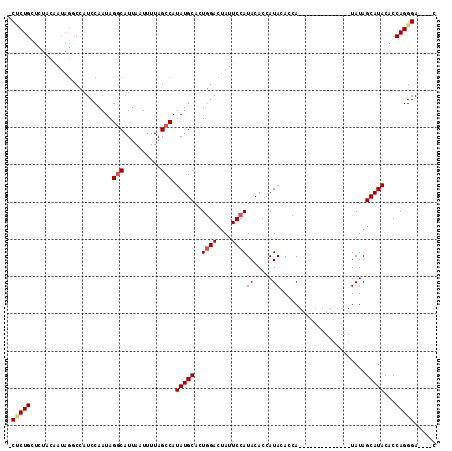
| Location | 27,300,517 – 27,300,614 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -16.84 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27300517 97 - 27905053 G----UCCCUGGUGUAUGCUAUA--------------UGGUGUAUAGUGUAUGGAAUAGUCCAGUGCAUAUGGCUAAAAUUAAUGCCUAUUGGAUGGACUAUUGUAGAGCAGAGG .----.(((((.(((((((((((--------------(...)))))))))))(.(((((((((.(.((...(((..........)))...)).)))))))))).)..).))).)) ( -29.30) >DroSec_CAF1 5521 110 - 1 G----UCCCUGGUGUAUGCUAUAUGCUAUAUGGUGUAUGGUGUAUGGUGUAUGGAAUAGUCCAGUGCAUAUGGCUAAAAUUAAUGACUAUUGGAUGGCCUAUUGUAGAGCAGGG- .----.(((((.(((((((((((((((((((...))))))))))))))))))(((....)))..((((...(((((.(((........)))...)))))...)))).).)))))- ( -34.90) >DroSim_CAF1 5942 96 - 1 G----UCCCUGGUGUAUGCUAUA--------------UGGUGUAUGGUGUAUGGAAUAGUCCAGUGCAUAUGGCUAAAAUUAAUGCCUAUUGGAUGGCCUAUUGUAGAGCAGAG- .----...(((.(((((((((((--------------(...)))))))))))((....((((((((.....(((..........)))))))))))..))........).)))..- ( -22.90) >DroYak_CAF1 1662 97 - 1 GUCGGUCCCUGGUGUAUGCUAUAU-------GUUGUAUGGUGUAUGGUGUAUGGAAUAGUCUAGUGCAUAUGGCUAAAAUUAAUGCCUAUUGGACUGUU----------CAGUG- .............((((((((((.-------.((....))..)))))))))).(((((((((((((.....(((..........)))))))))))))))----------)....- ( -27.80) >consensus G____UCCCUGGUGUAUGCUAUA______________UGGUGUAUGGUGUAUGGAAUAGUCCAGUGCAUAUGGCUAAAAUUAAUGCCUAUUGGAUGGCCUAUUGUAGAGCAGAG_ ........(((..(((((((((((.................)))))))))))......((((((((.....(((..........)))))))))))..............)))... (-16.84 = -17.03 + 0.19)


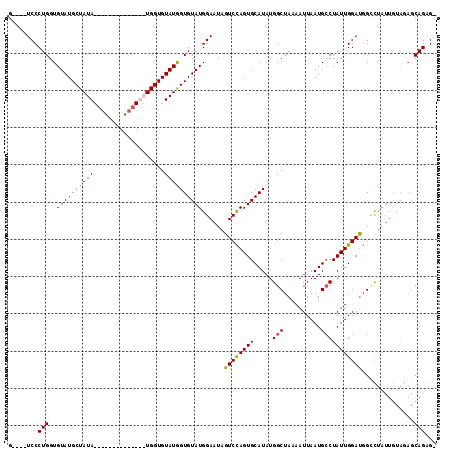
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:50:33 2006