
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,207,682 – 3,207,774 |
| Length | 92 |
| Max. P | 0.518948 |

| Location | 3,207,682 – 3,207,774 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 64.88 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -5.46 |
| Energy contribution | -6.72 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.19 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3207682 92 - 27905053 ACAGCCAAA---GGGGUGUGGAG--------UGCUGCGUAGCAAUGCUCAGCAUAAGCCACUUUUCACUCGGUUGGGGCAGUCAAUUGCAUGGAAAUUAAAUG-- .(((((...---(..(.((((.(--------(((((.(((....))).))))))...)))).)..)....)))))..((((....))))..............-- ( -29.40) >DroVir_CAF1 35887 88 - 1 ACAGUUCGACUUUAGUCGGGCUGUUUG----UAGGCCAU-----AGAUUG----GAAACACU--UAAAUUGAUU--GGCAUUUAAAUAAAUGCAUUUUUAAUGAC ((((((((((....)))))))))).((----(...(((.-----....))----)..)))..--...(((((..--.((((((....))))))....)))))... ( -23.60) >DroSec_CAF1 27076 92 - 1 ACAGCCAAA---GGGGUGUGGAG--------UGCUGCGUAGAAAUGCUCAGCAUAAGCCACUUUUCACUCGGUUGGGGCAGUCAAUUGCAUGGAAAUUAAAUG-- .(((((...---(..(.((((.(--------(((((.(((....))).))))))...)))).)..)....)))))..((((....))))..............-- ( -29.90) >DroSim_CAF1 33896 92 - 1 ACAGCCAAA---GGGGAGUGGAG--------UGCUGCGUAGAAAUGCUCAGCAUAAGCCACUUUUCACUCGGUUGGGGCAGUCAAUUGCAUGGAAAUUAAAUG-- .(((((...---(..((((((.(--------(((((.(((....))).))))))...))))))..)....)))))..((((....))))..............-- ( -34.40) >DroEre_CAF1 26559 91 - 1 ACAGCCAAA---GGGGUGUGGAG--------UGCUACGUAG-AAUGCUCAGCAUAAGCCACUUUUCACUCGGCUGGGGCAGUCAAUUGCAUGGAAAUUAAAUG-- .(((((...---(..(.((((.(--------((((..((..-...))..)))))...)))).)..)....)))))..((((....))))..............-- ( -24.50) >DroMoj_CAF1 34312 96 - 1 ACAGUUCGACUAUAGUCGGGCUGUUGGGCUCUUGGGCUCGGACAAGCCCA----GAGACACU--UUAAUGGCCU--GGUAUUUAGAUAAAUGCAUU-UGAAUGGC .(((((((((....)))))))))(..((((((((((((......))))))----.)))((..--....))))).--.)(((((((((......)))-)))))).. ( -33.00) >consensus ACAGCCAAA___GGGGUGUGGAG________UGCUGCGUAGAAAUGCUCAGCAUAAGCCACUUUUCACUCGGUUGGGGCAGUCAAUUGCAUGGAAAUUAAAUG__ ...(((..........................((((.(((....))).))))...((((...........))))..))).......................... ( -5.46 = -6.72 + 1.25)

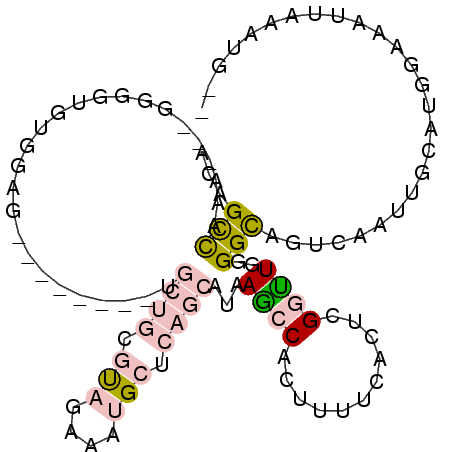
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:24 2006