
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,299,189 – 27,299,375 |
| Length | 186 |
| Max. P | 0.999966 |

| Location | 27,299,189 – 27,299,295 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -36.84 |
| Consensus MFE | -35.18 |
| Energy contribution | -35.27 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27299189 106 + 27905053 GGUCCUUGUCGACCAUUGGCAAUUACAUGCAGCUCACGUCCUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAG-------------- ((((......))))..((((.....(((((((....(((.((((((.((((((...........))))))))))))..))).....)))))))........)))).-------------- ( -33.12) >DroSec_CAF1 4202 106 + 1 GGUCCUUGUCGGCCAUUGGCAAUUACAUGCAGCUCACGUCCUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAG-------------- ..........((((((((((.....(((((((....(((.((((((.((((((...........))))))))))))..))).....))))))))))).))))))..-------------- ( -37.90) >DroSim_CAF1 4625 106 + 1 GGUCCUUGUCGGCCAUUGGCAAUUACAUGCAGCUCACGUCCUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAG-------------- ..........((((((((((.....(((((((....(((.((((((.((((((...........))))))))))))..))).....))))))))))).))))))..-------------- ( -37.90) >DroEre_CAF1 4376 106 + 1 GGUCCUUGUCGGCCAUUGGCAAUUACAUGCAGCUCACGUCCUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAG-------------- ..........((((((((((.....(((((((....(((.((((((.((((((...........))))))))))))..))).....))))))))))).))))))..-------------- ( -37.90) >DroYak_CAF1 306 106 + 1 GGUCCUUGUCGGCCAUUGGCAAUUACAUGCAGCUCACGUCCUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAG-------------- ..........((((((((((.....(((((((....(((.((((((.((((((...........))))))))))))..))).....))))))))))).))))))..-------------- ( -37.90) >DroAna_CAF1 5014 120 + 1 GGUCCUUGUCGGCCAUUGGCAAUUACAUGCAGCUCACGUCCUGUCAAAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGACCACAUGGUUAGAGUCGCCAAGACUG ((((......)))).(((((.....(((((((....(((.(((((((.(((((...........))))))))))))..))).....)))))))(((....))).......)))))..... ( -36.30) >consensus GGUCCUUGUCGGCCAUUGGCAAUUACAUGCAGCUCACGUCCUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAG______________ ..........((((((((((.....(((((((....(((.((((((.((((((...........))))))))))))..))).....))))))))))).))))))................ (-35.18 = -35.27 + 0.08)



| Location | 27,299,189 – 27,299,295 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -39.78 |
| Energy contribution | -39.87 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.98 |
| SVM RNA-class probability | 0.999966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27299189 106 - 27905053 --------------CUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAGGACGUGAGCUGCAUGUAAUUGCCAAUGGUCGACAAGGACC --------------.(((((((.((((((((((......((((.((((((((((((...........)))))).))))))..))))...)))))).....)))))))))))......... ( -39.20) >DroSec_CAF1 4202 106 - 1 --------------CUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAGGACGUGAGCUGCAUGUAAUUGCCAAUGGCCGACAAGGACC --------------.(((((((.((((((((((......((((.((((((((((((...........)))))).))))))..))))...)))))).....)))))))))))......... ( -41.90) >DroSim_CAF1 4625 106 - 1 --------------CUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAGGACGUGAGCUGCAUGUAAUUGCCAAUGGCCGACAAGGACC --------------.(((((((.((((((((((......((((.((((((((((((...........)))))).))))))..))))...)))))).....)))))))))))......... ( -41.90) >DroEre_CAF1 4376 106 - 1 --------------CUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAGGACGUGAGCUGCAUGUAAUUGCCAAUGGCCGACAAGGACC --------------.(((((((.((((((((((......((((.((((((((((((...........)))))).))))))..))))...)))))).....)))))))))))......... ( -41.90) >DroYak_CAF1 306 106 - 1 --------------CUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAGGACGUGAGCUGCAUGUAAUUGCCAAUGGCCGACAAGGACC --------------.(((((((.((((((((((......((((.((((((((((((...........)))))).))))))..))))...)))))).....)))))))))))......... ( -41.90) >DroAna_CAF1 5014 120 - 1 CAGUCUUGGCGACUCUAACCAUGUGGUCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUUUGACAGGACGUGAGCUGCAUGUAAUUGCCAAUGGCCGACAAGGACC ..((((((((((.....(((....)))((((((......((((.((((((((((((...........))))).)))))))..))))...))))))...))))))).)))........... ( -38.70) >consensus ______________CUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAGGACGUGAGCUGCAUGUAAUUGCCAAUGGCCGACAAGGACC ...............(((((((.((((((((((......((((.((((((((((((...........)))))).))))))..))))...)))))).....)))))))))))......... (-39.78 = -39.87 + 0.09)



| Location | 27,299,229 – 27,299,335 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 99.06 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -31.92 |
| Energy contribution | -31.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27299229 106 + 27905053 CUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGGAAAAAAU (((((((((((((...........))))))...((((..(((..((((((.(((((....)))))))))))..).))..)))).......))).))))........ ( -32.00) >DroSec_CAF1 4242 106 + 1 CUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGGAAAAAAU (((((((((((((...........))))))...((((..(((..((((((.(((((....)))))))))))..).))..)))).......))).))))........ ( -32.00) >DroSim_CAF1 4665 106 + 1 CUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGGAAAAAAU (((((((((((((...........))))))...((((..(((..((((((.(((((....)))))))))))..).))..)))).......))).))))........ ( -32.00) >DroEre_CAF1 4416 106 + 1 CUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGAAAAAAAU (((((((((((((...........))))))...((((..(((..((((((.(((((....)))))))))))..).))..)))).......))).))))........ ( -31.80) >DroYak_CAF1 346 106 + 1 CUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGAAAGAAAU (((((((((((((...........))))))...((((..(((..((((((.(((((....)))))))))))..).))..)))).......))).))))........ ( -31.80) >consensus CUGUCAGAGUUGAAAGAUGACAAAUCAACUUGACAGCCGCGCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGGAAAAAAU (((((((((((((...........))))))...((((..(((..((((((.(((((....)))))))))))..).))..)))).......))).))))........ (-31.92 = -31.92 + -0.00)

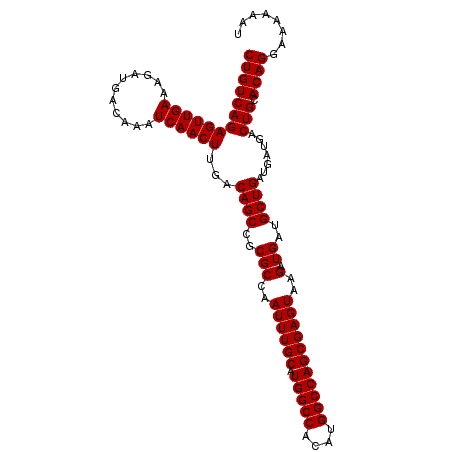

| Location | 27,299,229 – 27,299,335 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 99.06 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -31.92 |
| Energy contribution | -31.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27299229 106 - 27905053 AUUUUUUCCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAG ........((((((.((........))............(((((((....))))).))......))))))((((((((((((...........)))))).)))))) ( -32.00) >DroSec_CAF1 4242 106 - 1 AUUUUUUCCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAG ........((((((.((........))............(((((((....))))).))......))))))((((((((((((...........)))))).)))))) ( -32.00) >DroSim_CAF1 4665 106 - 1 AUUUUUUCCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAG ........((((((.((........))............(((((((....))))).))......))))))((((((((((((...........)))))).)))))) ( -32.00) >DroEre_CAF1 4416 106 - 1 AUUUUUUUCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAG ........((((((.((........))............(((((((....))))).))......))))))((((((((((((...........)))))).)))))) ( -31.80) >DroYak_CAF1 346 106 - 1 AUUUCUUUCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAG ........((((((.((........))............(((((((....))))).))......))))))((((((((((((...........)))))).)))))) ( -31.80) >consensus AUUUUUUCCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGCGCGGCUGUCAAGUUGAUUUGUCAUCUUUCAACUCUGACAG ........((((((.((........))............(((((((....))))).))......))))))((((((((((((...........)))))).)))))) (-31.92 = -31.92 + -0.00)



| Location | 27,299,269 – 27,299,375 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -21.86 |
| Energy contribution | -21.66 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27299269 106 + 27905053 GCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGGAAAAAAUUAGAUCAUCAACUUGAUCAAUAACUCAAAUAGUAUUGUUU ....((((((.(((((....))))))))))).(((..((((((.(((((((((.............))).))))))..((((........)))).))))))..))) ( -29.22) >DroSec_CAF1 4282 106 + 1 GCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGGAAAAAAUUAGAUCAUCAACAUGAUCAAUAACUCAAAUGGUAUUGUUU ((((((((((.(((((....)))))))))))..(((.(((.(((((((.....................))))))).))).)))...........))))....... ( -31.80) >DroSim_CAF1 4705 106 + 1 GCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGGAAAAAAUUAGAUCAUCAACUUGAUCAAUAACUCAAAUGGUAUUGUUU ((((((((((.(((((....)))))))))))..((((((.(((((.....((....))......)))))))))))...((((........)))).))))....... ( -30.60) >DroEre_CAF1 4456 106 + 1 GCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGAAAAAAAUAUAACCAUCAACUUGAUUAAUAACUCAAUUUUUUAUGUUU ....((((((.(((((....)))))))))))(((.((((((((.((.......))))).............))))).))).......................... ( -20.11) >DroYak_CAF1 386 106 + 1 GCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGAAAGAAAUAUUAUCGUCAACUUGAUUAAUAAUUCAAAUUUAAUUGUUC ....((((((.(((((....)))))))))))..((..((..(((((((((((........)).....)))))))))..((((........)))).....))..)). ( -24.10) >consensus GCCAAUUUGCAUGGCCACAUGGCCAGCGAGUAAGAUGAUGCUGAUGAUGACUGCACAGGAAAAAAUUAGAUCAUCAACUUGAUCAAUAACUCAAAUGGUAUUGUUU ....((((((.(((((....)))))))))))..(((.(.(.(((((((.....................))))))).).).)))...................... (-21.86 = -21.66 + -0.20)



| Location | 27,299,269 – 27,299,375 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -23.36 |
| Energy contribution | -24.40 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27299269 106 - 27905053 AAACAAUACUAUUUGAGUUAUUGAUCAAGUUGAUGAUCUAAUUUUUUCCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGC ...((((.......((((...((((...(((((((((((.((........))..))..)))))))))))))....))))(((((((....))))).))..)))).. ( -29.00) >DroSec_CAF1 4282 106 - 1 AAACAAUACCAUUUGAGUUAUUGAUCAUGUUGAUGAUCUAAUUUUUUCCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGC ........(((...((((....(((.(((((((((((((.((........))..))..))))))))))).)))..))))(((((((....))))).))....))). ( -32.70) >DroSim_CAF1 4705 106 - 1 AAACAAUACCAUUUGAGUUAUUGAUCAAGUUGAUGAUCUAAUUUUUUCCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGC ........(((...((((...((((...(((((((((((.((........))..))..)))))))))))))....))))(((((((....))))).))....))). ( -29.20) >DroEre_CAF1 4456 106 - 1 AAACAUAAAAAAUUGAGUUAUUAAUCAAGUUGAUGGUUAUAUUUUUUUCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGC ..............((((..........(((((((((.((...............)).)))))))))........))))(((((((....))))).))........ ( -23.23) >DroYak_CAF1 386 106 - 1 GAACAAUUAAAUUUGAAUUAUUAAUCAAGUUGACGAUAAUAUUUCUUUCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGC ...(((((....((((........))))(((((.(((.((....(.....)....)).))).)))))............(((((((....))))).)).))))).. ( -20.60) >consensus AAACAAUACAAUUUGAGUUAUUGAUCAAGUUGAUGAUCUAAUUUUUUCCUGUGCAGUCAUCAUCAGCAUCAUCUUACUCGCUGGCCAUGUGGCCAUGCAAAUUGGC ...((((.......((((...((((...(((((((((.....................)))))))))))))....))))(((((((....))))).))..)))).. (-23.36 = -24.40 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:50:27 2006