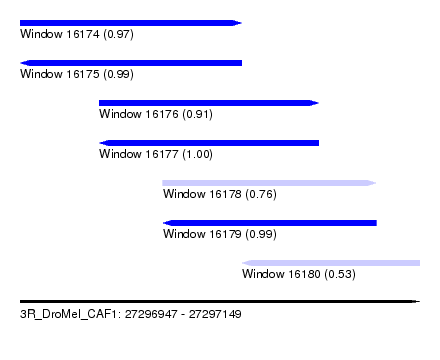
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,296,947 – 27,297,149 |
| Length | 202 |
| Max. P | 0.999658 |

| Location | 27,296,947 – 27,297,059 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.73 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -19.89 |
| Energy contribution | -24.22 |
| Covariance contribution | 4.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27296947 112 + 27905053 CUCAGUUCCCUGUGUGUUGUAAGCUGAAAACAAUUUACGGGAACCAGCCU---UCUGCAU-----UCUGCAUUCUGGAUUCUGGCCAUUGGCCAUUCGCCAUUGGCCAGCUGUUUGGCCA ....((((((.(((.(((((.........))))).)))))))))..(((.---..(((..-----...)))....((((.(((((((.((((.....)))).)))))))..))))))).. ( -40.80) >DroSec_CAF1 2016 105 + 1 CUCUGUUCCCUGUGUGUUGUAAGCUGAAAACAAUUUACGGGAACCAGCCU---UCUGCAU-----UCUGCAUUCUGGAUUCUGGCCAUUGG-------CCAUUGGCCAGCUGUUUGGCCA ....((((((.(((.(((((.........))))).)))))))))..(((.---..(((..-----...)))....((((.(((((((....-------....)))))))..))))))).. ( -33.20) >DroSim_CAF1 2424 112 + 1 CUCUGUUCCCUGUGUGUUGUAAGCUGAAAACAAUUUACGGGAACCAGCCU---UCUGCAU-----UCUGGAUUCUGGAUUCUGGCCAUUGGCCAUUGGCCAUUGGCCAGCUGUUUGGCCA ....((((((.(((.(((((.........))))).)))))))))..(((.---...(((.-----((..(...)..))..(((((((.(((((...))))).))))))).)))..))).. ( -42.50) >DroEre_CAF1 2148 98 + 1 CUCUGUUCCCUGUGUGUUGUAAGCUGAAAACCAUUUACGGGAACCAGCCU---UCAGCCU-----UCUGCAUUCUGGAUUCUGGCCAU--------------UGGCCAGCUGUUUGGCCA ....((((((.(((.(((..........))))))....))))))..(((.---.((((..-----(((.......)))...(((((..--------------.)))))))))...))).. ( -27.80) >DroAna_CAF1 3121 100 + 1 CU----UCUGGGUCCGAGGAAAACUGAAAACAAUUUAUGGGAACCGUACGGCCGGUGCAACUGCAACUGCAACUU----------------CAACUGGCCAAGAACCAGCUGUUUGGCCA ..----....((.(((((.(...(((.........(((((...))))).(((((((.....(((....)))....----------------..)))))))......))).).))))))). ( -27.10) >consensus CUCUGUUCCCUGUGUGUUGUAAGCUGAAAACAAUUUACGGGAACCAGCCU___UCUGCAU_____UCUGCAUUCUGGAUUCUGGCCAUUGGC_A_U_GCCAUUGGCCAGCUGUUUGGCCA ....((((((.(((.(((((.........))))).)))))))))..(((.......(((........)))..........(((((((.((((.....)))).)))))))......))).. (-19.89 = -24.22 + 4.33)



| Location | 27,296,947 – 27,297,059 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.73 |
| Mean single sequence MFE | -34.41 |
| Consensus MFE | -20.33 |
| Energy contribution | -24.82 |
| Covariance contribution | 4.49 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27296947 112 - 27905053 UGGCCAAACAGCUGGCCAAUGGCGAAUGGCCAAUGGCCAGAAUCCAGAAUGCAGA-----AUGCAGA---AGGCUGGUUCCCGUAAAUUGUUUUCAGCUUACAACACACAGGGAACUGAG .((((......(((((((.((((.....)))).))))))).........(((...-----..)))..---.))))(((((((((...((((.........))))...)).)))))))... ( -41.10) >DroSec_CAF1 2016 105 - 1 UGGCCAAACAGCUGGCCAAUGG-------CCAAUGGCCAGAAUCCAGAAUGCAGA-----AUGCAGA---AGGCUGGUUCCCGUAAAUUGUUUUCAGCUUACAACACACAGGGAACAGAG .((((......((((....(((-------((...)))))....))))..(((...-----..)))..---.)))).((((((((...((((.........))))...)).)))))).... ( -32.90) >DroSim_CAF1 2424 112 - 1 UGGCCAAACAGCUGGCCAAUGGCCAAUGGCCAAUGGCCAGAAUCCAGAAUCCAGA-----AUGCAGA---AGGCUGGUUCCCGUAAAUUGUUUUCAGCUUACAACACACAGGGAACAGAG .((((......(((((((.(((((...))))).)))))))......(....)...-----.......---.)))).((((((((...((((.........))))...)).)))))).... ( -37.40) >DroEre_CAF1 2148 98 - 1 UGGCCAAACAGCUGGCCA--------------AUGGCCAGAAUCCAGAAUGCAGA-----AGGCUGA---AGGCUGGUUCCCGUAAAUGGUUUUCAGCUUACAACACACAGGGAACAGAG .((((......((((((.--------------..))))))....(((..(.....-----)..))).---.)))).((((((.......((.........))........)))))).... ( -27.36) >DroAna_CAF1 3121 100 - 1 UGGCCAAACAGCUGGUUCUUGGCCAGUUG----------------AAGUUGCAGUUGCAGUUGCACCGGCCGUACGGUUCCCAUAAAUUGUUUUCAGUUUUCCUCGGACCCAGA----AG .((((...((((((((.....))))))))----------------..(.(((((......))))).)))))....((.(((...((((((....)))))).....))).))...----.. ( -33.30) >consensus UGGCCAAACAGCUGGCCAAUGGC_A_U_GCCAAUGGCCAGAAUCCAGAAUGCAGA_____AUGCAGA___AGGCUGGUUCCCGUAAAUUGUUUUCAGCUUACAACACACAGGGAACAGAG .((((......(((((((.((((.....)))).))))))).........((((........))))......)))).((((((((((.(((....))).))))........)))))).... (-20.33 = -24.82 + 4.49)



| Location | 27,296,987 – 27,297,098 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.02 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -22.45 |
| Energy contribution | -25.34 |
| Covariance contribution | 2.89 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27296987 111 + 27905053 GAACCAGCCU---UCUGCAU-----UCUGCAUUCUGGAUUCUGGCCAUUGGCCAUUCGCCAUUGGCCAGCUGUUUGGCCAAGUGCAUUGACCGCCUGCAAAUUGCACUC-UGUAGCCAGU ...((((...---..(((..-----...)))..))))...(((((((.((((.....)))).)))))))....((((((((((((((((........)))..)))))).-))..))))). ( -39.00) >DroSec_CAF1 2056 104 + 1 GAACCAGCCU---UCUGCAU-----UCUGCAUUCUGGAUUCUGGCCAUUGG-------CCAUUGGCCAGCUGUUUGGCCAAGUGCAUUGACCGCCUGCAAAUUGCACUC-UGCAGCCAGU ...((((...---..(((..-----...)))..))))...(((((((((((-------((...)))))).))....((..(((((((((........)))..)))))).-.)).))))). ( -32.30) >DroSim_CAF1 2464 111 + 1 GAACCAGCCU---UCUGCAU-----UCUGGAUUCUGGAUUCUGGCCAUUGGCCAUUGGCCAUUGGCCAGCUGUUUGGCCAAGUGCAUUGACCGCCUGCAAAUUGCACUC-UGCAGCCAGU ...((((..(---((.....-----...)))..))))...(((((((.(((((...))))).)))))))....((((((((((((((((........)))..)))))).-))..))))). ( -41.60) >DroEre_CAF1 2188 97 + 1 GAACCAGCCU---UCAGCCU-----UCUGCAUUCUGGAUUCUGGCCAU--------------UGGCCAGCUGUUUGGCCAAGUGCAUUGACCGCCUGCAAAUUGCACUC-UGCAGCCAGU ...((((...---.(((...-----.)))....))))...(((((..(--------------(((((((....))))))))((((((((........)))..)))))..-....))))). ( -30.90) >DroAna_CAF1 3157 104 + 1 GAACCGUACGGCCGGUGCAACUGCAACUGCAACUU----------------CAACUGGCCAAGAACCAGCUGUUUGGCCAAGUGCAAUGACCGUCUGCAAAUUGCACUCUUGCAGCCAUC .........(((((((.....(((....)))....----------------..)))))))........(((((..((...((((((((............)))))))))).))))).... ( -31.40) >consensus GAACCAGCCU___UCUGCAU_____UCUGCAUUCUGGAUUCUGGCCAUUGGC_A_U_GCCAUUGGCCAGCUGUUUGGCCAAGUGCAUUGACCGCCUGCAAAUUGCACUC_UGCAGCCAGU ....((((........(((........)))...........((((((.((((.....)))).))))))))))..(((((((((((((((........)))..))))))..))..)))).. (-22.45 = -25.34 + 2.89)



| Location | 27,296,987 – 27,297,098 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.02 |
| Mean single sequence MFE | -42.84 |
| Consensus MFE | -27.37 |
| Energy contribution | -30.42 |
| Covariance contribution | 3.05 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27296987 111 - 27905053 ACUGGCUACA-GAGUGCAAUUUGCAGGCGGUCAAUGCACUUGGCCAAACAGCUGGCCAAUGGCGAAUGGCCAAUGGCCAGAAUCCAGAAUGCAGA-----AUGCAGA---AGGCUGGUUC ((..(((...-...((((.((((((..(((((((.....))))))......(((((((.((((.....)))).)))))))......)..))))))-----.))))..---.)))..)).. ( -47.10) >DroSec_CAF1 2056 104 - 1 ACUGGCUGCA-GAGUGCAAUUUGCAGGCGGUCAAUGCACUUGGCCAAACAGCUGGCCAAUGG-------CCAAUGGCCAGAAUCCAGAAUGCAGA-----AUGCAGA---AGGCUGGUUC ((((.(((((-((......))))))).))))........(((((((......)))))))(((-------((...)))))(((.((((..(((...-----..)))..---...))))))) ( -41.00) >DroSim_CAF1 2464 111 - 1 ACUGGCUGCA-GAGUGCAAUUUGCAGGCGGUCAAUGCACUUGGCCAAACAGCUGGCCAAUGGCCAAUGGCCAAUGGCCAGAAUCCAGAAUCCAGA-----AUGCAGA---AGGCUGGUUC .(((((((((-((......)))))))..((((((.....))))))......(((((((.(((((...))))).)))))))...))))...((((.-----.(.....---)..))))... ( -47.20) >DroEre_CAF1 2188 97 - 1 ACUGGCUGCA-GAGUGCAAUUUGCAGGCGGUCAAUGCACUUGGCCAAACAGCUGGCCA--------------AUGGCCAGAAUCCAGAAUGCAGA-----AGGCUGA---AGGCUGGUUC ((((.(((((-((......))))))).))))....((..(((((((......))))))--------------)..))..(((.((((....(((.-----...))).---...))))))) ( -36.60) >DroAna_CAF1 3157 104 - 1 GAUGGCUGCAAGAGUGCAAUUUGCAGACGGUCAUUGCACUUGGCCAAACAGCUGGUUCUUGGCCAGUUG----------------AAGUUGCAGUUGCAGUUGCACCGGCCGUACGGUUC .(((((((...((.(((((((.((((.((((((.......)))))...((((((((.....))))))))----------------..)))))))))))).))....)))))))....... ( -42.30) >consensus ACUGGCUGCA_GAGUGCAAUUUGCAGGCGGUCAAUGCACUUGGCCAAACAGCUGGCCAAUGGC_A_U_GCCAAUGGCCAGAAUCCAGAAUGCAGA_____AUGCAGA___AGGCUGGUUC ..(((((....(((((((..(((....)))....))))))))))))..((((((((((.((((.....)))).)))))...........((((........))))......))))).... (-27.37 = -30.42 + 3.05)


| Location | 27,297,019 – 27,297,127 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.38 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -27.07 |
| Energy contribution | -29.36 |
| Covariance contribution | 2.29 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27297019 108 + 27905053 CUGGCCAUUGGCCAUUCGCCAUUGGCCAGCUGUUUGGCCAAGUGCAUUGACCGCCUGCAAAUUGCACUC-UGUAGCCAGUUCGCUGCACAAAUUGAUUUGAAUGAAAGC----------- (((((((.((((.....)))).)))))))(.(((((....(((((((((........)))..)))))).-((((((......))))))))))).)..............----------- ( -35.70) >DroSec_CAF1 2088 101 + 1 CUGGCCAUUGG-------CCAUUGGCCAGCUGUUUGGCCAAGUGCAUUGACCGCCUGCAAAUUGCACUC-UGCAGCCAGUUCGCUGCACAAAUUGAUUUGAAUGAAAGC----------- .(((((...))-------)))((((((((....))))))))(((((.(((.(..(((((..........-)))))...).))).)))))....................----------- ( -32.70) >DroSim_CAF1 2496 108 + 1 CUGGCCAUUGGCCAUUGGCCAUUGGCCAGCUGUUUGGCCAAGUGCAUUGACCGCCUGCAAAUUGCACUC-UGCAGCCAGUUCGCUGCACAAAUUGAUUUGAAUGAAAGC----------- (((((((.(((((...))))).)))))))(.(((((....(((((((((........)))..)))))).-((((((......))))))))))).)..............----------- ( -40.00) >DroEre_CAF1 2220 94 + 1 CUGGCCAU--------------UGGCCAGCUGUUUGGCCAAGUGCAUUGACCGCCUGCAAAUUGCACUC-UGCAGCCAGUUCGCUGCACAAAUUGAUUUGAAUGAAAGC----------- ((((((..--------------.))))))(.(((((....(((((((((........)))..)))))).-((((((......))))))))))).)..............----------- ( -27.00) >DroAna_CAF1 3192 109 + 1 -----------CAACUGGCCAAGAACCAGCUGUUUGGCCAAGUGCAAUGACCGUCUGCAAAUUGCACUCUUGCAGCCAUCUCGCUGCACAAAUUGAUUUGAAUGAAAGCUGCCCAGGUCA -----------....((((((((.........))))))))((((((((............))))))))..((((((......)))))).....(((((((.............))))))) ( -30.82) >consensus CUGGCCAUUGGC_A_U_GCCAUUGGCCAGCUGUUUGGCCAAGUGCAUUGACCGCCUGCAAAUUGCACUC_UGCAGCCAGUUCGCUGCACAAAUUGAUUUGAAUGAAAGC___________ (((((((.((((.....)))).)))))))(.(((((....(((((((((........)))..))))))..((((((......))))))))))).)......................... (-27.07 = -29.36 + 2.29)


| Location | 27,297,019 – 27,297,127 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.38 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -29.61 |
| Energy contribution | -32.46 |
| Covariance contribution | 2.85 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27297019 108 - 27905053 -----------GCUUUCAUUCAAAUCAAUUUGUGCAGCGAACUGGCUACA-GAGUGCAAUUUGCAGGCGGUCAAUGCACUUGGCCAAACAGCUGGCCAAUGGCGAAUGGCCAAUGGCCAG -----------(((......((((....))))(((((((..(((....))-)..)))....)))))))((((((.....))))))......(((((((.((((.....)))).))))))) ( -37.30) >DroSec_CAF1 2088 101 - 1 -----------GCUUUCAUUCAAAUCAAUUUGUGCAGCGAACUGGCUGCA-GAGUGCAAUUUGCAGGCGGUCAAUGCACUUGGCCAAACAGCUGGCCAAUGG-------CCAAUGGCCAG -----------....................(((((..((.(((.(((((-((......))))))).)))))..)))))(((((((......)))))))(((-------((...))))). ( -41.00) >DroSim_CAF1 2496 108 - 1 -----------GCUUUCAUUCAAAUCAAUUUGUGCAGCGAACUGGCUGCA-GAGUGCAAUUUGCAGGCGGUCAAUGCACUUGGCCAAACAGCUGGCCAAUGGCCAAUGGCCAAUGGCCAG -----------....................(((((..((.(((.(((((-((......))))))).)))))..)))))............(((((((.(((((...))))).))))))) ( -44.80) >DroEre_CAF1 2220 94 - 1 -----------GCUUUCAUUCAAAUCAAUUUGUGCAGCGAACUGGCUGCA-GAGUGCAAUUUGCAGGCGGUCAAUGCACUUGGCCAAACAGCUGGCCA--------------AUGGCCAG -----------(((.................(((((..((.(((.(((((-((......))))))).)))))..)))))(((((((......))))))--------------).)))... ( -35.50) >DroAna_CAF1 3192 109 - 1 UGACCUGGGCAGCUUUCAUUCAAAUCAAUUUGUGCAGCGAGAUGGCUGCAAGAGUGCAAUUUGCAGACGGUCAUUGCACUUGGCCAAACAGCUGGUUCUUGGCCAGUUG----------- .......(((......................((((((......)))))).(((((((((..((.....)).))))))))).)))...((((((((.....))))))))----------- ( -36.30) >consensus ___________GCUUUCAUUCAAAUCAAUUUGUGCAGCGAACUGGCUGCA_GAGUGCAAUUUGCAGGCGGUCAAUGCACUUGGCCAAACAGCUGGCCAAUGGC_A_U_GCCAAUGGCCAG ...........((((.....(((((..((((.((((((......)))))).))))...))))).))))((((((.....))))))......(((((((.((((.....)))).))))))) (-29.61 = -32.46 + 2.85)

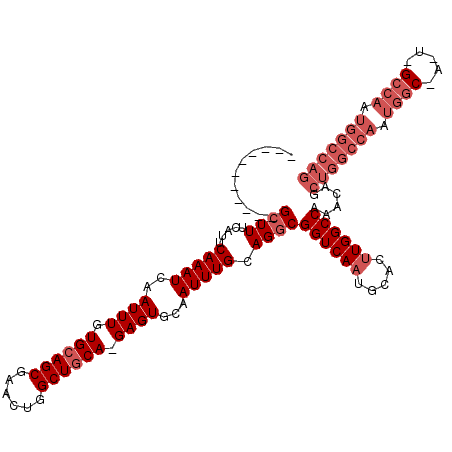

| Location | 27,297,059 – 27,297,149 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -24.17 |
| Energy contribution | -24.43 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.98 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27297059 90 - 27905053 CAGCACAAUCAGCAUUCCCUCCGCUUUCAUUCAAAUCAAUUUGUGCAGCGAACUGGCUACAGAGUGCAAUUUGCAGGCGGUCAAUGCACU ...........(((((....(((((......((((....))))(((((((..(((....)))..)))....)))))))))..)))))... ( -21.10) >DroSec_CAF1 2121 90 - 1 CAGCACAAUCAGCAUUCCCUCCGCUUUCAUUCAAAUCAAUUUGUGCAGCGAACUGGCUGCAGAGUGCAAUUUGCAGGCGGUCAAUGCACU ...........(((((....((((((.....(((((....((.((((((......)))))).))....))))).))))))..)))))... ( -25.80) >DroSim_CAF1 2536 90 - 1 CAGCACAAUCAGCAUUCCCUCCGCUUUCAUUCAAAUCAAUUUGUGCAGCGAACUGGCUGCAGAGUGCAAUUUGCAGGCGGUCAAUGCACU ...........(((((....((((((.....(((((....((.((((((......)))))).))....))))).))))))..)))))... ( -25.80) >DroEre_CAF1 2246 90 - 1 CAGCACAAUCAGCAUUCCCUCCGCUUUCAUUCAAAUCAAUUUGUGCAGCGAACUGGCUGCAGAGUGCAAUUUGCAGGCGGUCAAUGCACU ...........(((((....((((((.....(((((....((.((((((......)))))).))....))))).))))))..)))))... ( -25.80) >consensus CAGCACAAUCAGCAUUCCCUCCGCUUUCAUUCAAAUCAAUUUGUGCAGCGAACUGGCUGCAGAGUGCAAUUUGCAGGCGGUCAAUGCACU ...........(((((....((((((.....(((((....((.((((((......)))))).))....))))).))))))..)))))... (-24.17 = -24.43 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:50:21 2006