
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,254,429 – 27,254,526 |
| Length | 97 |
| Max. P | 0.811136 |

| Location | 27,254,429 – 27,254,526 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -21.94 |
| Energy contribution | -23.17 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27254429 97 - 27905053 GCAUGACAGCUAUGCGGUGUGGGUUGGAAAACAAGUG------------AAACUGGUCGAGAUUGGUCUUGAUUGGUUCUC-----GGUUUCCGUUCCCC-----CCACCUUUUCGCCG (((((.....)))))((((.(((.((((((....(.(------------((.(..(((((((....)))))))..)))).)-----..))))))...)))-----.))))......... ( -35.60) >DroSec_CAF1 27626 96 - 1 GCAUGACAGCUAUGCGGUGUGGGUUGGAAAACAAGUG------------AGACUGGUCGAGAUUGGUCUUGAUUGGUUCUC-----GGUUUCCGUUCC-C-----CCACCUUUUCGCCG (((((.....)))))((((.(((.((((((.....((------------((((..(((((((....)))))))..).))))-----).))))))..))-)-----.))))......... ( -38.20) >DroSim_CAF1 27362 96 - 1 GCAUGACAGCUAUGCGGUGUGGGUUGGAAAACAAGUG------------AGACUGGUCGAGAUUGGUCUUGAUUGGUUCUC-----GGUUUCCGUUCC-C-----CCACCUUUUCGCCG (((((.....)))))((((.(((.((((((.....((------------((((..(((((((....)))))))..).))))-----).))))))..))-)-----.))))......... ( -38.20) >DroEre_CAF1 28164 97 - 1 GCAUGACAGCUAUGCGGUGUGGGUUGGAAAACAAGUG------------AGACUGGUCGAGAUUGGUCUUGAUUGGUUCUC-----GGUUGCUGUUCCCC-----CCACCUUAUCGCCA (((((.....)))))((((.(((..((((..(((.((------------((((..(((((((....)))))))..).))))-----).)))...))))))-----)))))......... ( -40.90) >DroYak_CAF1 29436 107 - 1 GCAUGACAGCAAUGCGGUGUGGGUUGGAAAACAAGUG------------AGACUGGUCGAGAUUGCUCUUGAUUGGUUCCCUCUGCUGUUGCUGUUUCCCCGUUUCCACCUUAUCGCCA ((((.......))))((((.((((.(((((...((.(------------..((..(((((((....)))))))..))..)))..((....))..........)))))))))...)))). ( -35.20) >DroAna_CAF1 24931 111 - 1 GCAUGACAGCAACGCGGUAUCAGGAGGAAAACAAGGAGACUGUGCUUGUUGACUGGUCGAGAUUGGACUUGAUUGGGUUU------GGUUUUUGUCUGUCC--UUCCCCCCGGACAGGA ......((((((((((((.((.(........)...)).)))))).)))))).(..((((((......))))))..)....------........(((((((--........))))))). ( -33.20) >consensus GCAUGACAGCUAUGCGGUGUGGGUUGGAAAACAAGUG____________AGACUGGUCGAGAUUGGUCUUGAUUGGUUCUC_____GGUUUCCGUUCCCC_____CCACCUUUUCGCCA (((((.....)))))((((.((((.((.......................(((..(((((((....)))))))..)))........((........)).......))))))...)))). (-21.94 = -23.17 + 1.22)

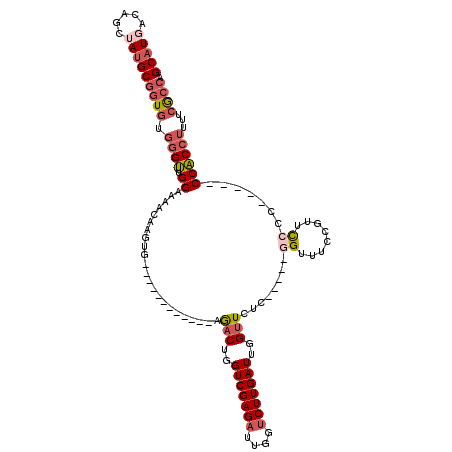

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:57 2006