
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,248,610 – 27,248,707 |
| Length | 97 |
| Max. P | 0.973000 |

| Location | 27,248,610 – 27,248,707 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.52 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -12.35 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
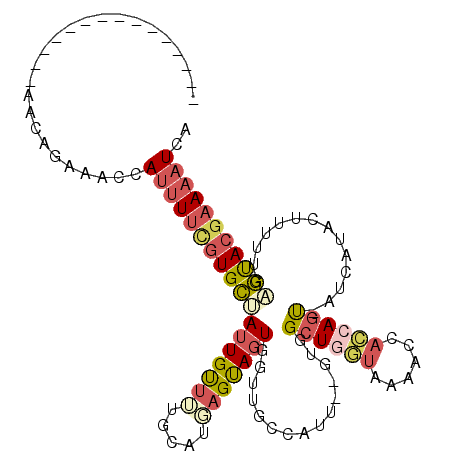
>3R_DroMel_CAF1 27248610 97 + 27905053 ---------------AAUAGAAACCAUUUUCGUGCAAUUGUUUUGCAUGAGUAGUGGUUGCAAUU--GUGGCUGGUAAAACCACCAGU-AUCAUACUUUUUAAGUACGAAAAUCA ---------------....(.((((((((((((((((.....))))))))).))))))).)....--...((((((......))))))-.((.((((.....)))).))...... ( -28.50) >DroSec_CAF1 21803 93 + 1 ---------------AAUAGAAACCAUUUUCGUGCUAUUGUUUUGCAUGAGUAGUGGCUGCCAUU--GUGGCUGGUAAAACCACCAAC-----CACUUUUUGAGUACGAAAAUCA ---------------..........(((((((((((..........(((.((((...))))))).--((((.((((......)))).)-----)))......))))))))))).. ( -25.00) >DroSim_CAF1 21907 97 + 1 ---------------AAUAGAAACCAUUUUCGUGCUAUUGUUUUGCAUGAGUAGUGGUUGCCAUU--GUGGCUGGUAAAACCACCAGU-AUCAUACUUUUUGAGUACGAAAAUCA ---------------....((((((((((((((((.........))))))).)))))))....((--(((((((((......))))))-.(((.......))).)))))...)). ( -26.10) >DroEre_CAF1 22002 97 + 1 ---------------AACAGAAACCAUUUUCCUGCAAUUGCUUUGCAUCAGUAGUGGUUGCCAUU--GUGGCUAGUAAAACUACCAGU-AUCAUACUUUUUGAGUACGAAAAUUA ---------------.((((.(((((((....(((((.....))))).....)))))))....))--))((.(((.....)))))...-.((.(((((...))))).))...... ( -18.40) >DroYak_CAF1 22541 95 + 1 ---------------AACAGAAACCAUUUUCGUGCUAUUGUUUUGCAUCAGCAGUGGUUGCCAUU--GCGGCUGUUAGAACAUUCAGU-AUCAUCCUAUUUGAGUACAAAAAU-- ---------------....((((....))))((((...(((((((((.(.(((((((...)))))--)).).)).)))))))......-.(((.......)))))))......-- ( -20.40) >DroPer_CAF1 28302 110 + 1 GCUUAUUGUUUUGUCAACAGAGUCU-UCUCUGUGGCAGUGCGAUACAGUGGCAGUCAUGGCCAUAAGGAGGCUAAUGAAACCAUCAGCCACCAUAGAAAUUGGCCACGAAA---- ...((((((.((((((.(((((...-.))))))))))).))))))..(((((..((((((((....)).((((.(((....))).)))).)))).)).....)))))....---- ( -40.70) >consensus _______________AACAGAAACCAUUUUCGUGCUAUUGUUUUGCAUGAGUAGUGGUUGCCAUU__GUGGCUGGUAAAACCACCAGU_AUCAUACUUUUUGAGUACGAAAAUCA .........................((((((((((((((((((.....)))))))...............((((((......))))))..............))))))))))).. (-12.35 = -13.32 + 0.96)

| Location | 27,248,610 – 27,248,707 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.52 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -11.12 |
| Energy contribution | -11.57 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27248610 97 - 27905053 UGAUUUUCGUACUUAAAAAGUAUGAU-ACUGGUGGUUUUACCAGCCAC--AAUUGCAACCACUACUCAUGCAAAACAAUUGCACGAAAAUGGUUUCUAUU--------------- ......(((((((.....))))))).-....((((((.....))))))--....(.(((((....((.(((((.....))))).))...))))).)....--------------- ( -21.70) >DroSec_CAF1 21803 93 - 1 UGAUUUUCGUACUCAAAAAGUG-----GUUGGUGGUUUUACCAGCCAC--AAUGGCAGCCACUACUCAUGCAAAACAAUAGCACGAAAAUGGUUUCUAUU--------------- ...................(((-----(((((((....))))))))))--(((((.(((((....((.(((.........))).))...))))).)))))--------------- ( -25.70) >DroSim_CAF1 21907 97 - 1 UGAUUUUCGUACUCAAAAAGUAUGAU-ACUGGUGGUUUUACCAGCCAC--AAUGGCAACCACUACUCAUGCAAAACAAUAGCACGAAAAUGGUUUCUAUU--------------- ..((((((((.((......((((((.-..((((((((......(((..--...))))))))))).))))))........)).))))))))..........--------------- ( -24.64) >DroEre_CAF1 22002 97 - 1 UAAUUUUCGUACUCAAAAAGUAUGAU-ACUGGUAGUUUUACUAGCCAC--AAUGGCAACCACUACUGAUGCAAAGCAAUUGCAGGAAAAUGGUUUCUGUU--------------- ......(((((((.....))))))).-.((((((....))))))....--(((((.(((((.......(((((.....)))))......))))).)))))--------------- ( -20.42) >DroYak_CAF1 22541 95 - 1 --AUUUUUGUACUCAAAUAGGAUGAU-ACUGAAUGUUCUAACAGCCGC--AAUGGCAACCACUGCUGAUGCAAAACAAUAGCACGAAAAUGGUUUCUGUU--------------- --..(((((((.(((..((((((...-.......))))))......((--(.(((...))).)))))))))))))....((((.((((....))))))))--------------- ( -15.60) >DroPer_CAF1 28302 110 - 1 ----UUUCGUGGCCAAUUUCUAUGGUGGCUGAUGGUUUCAUUAGCCUCCUUAUGGCCAUGACUGCCACUGUAUCGCACUGCCACAGAGA-AGACUCUGUUGACAAAACAAUAAGC ----..(((((((((.....((.((.((((((((....)))))))).)).))))))))))).(((.........))).(((.((((((.-...)))))).).))........... ( -35.80) >consensus UGAUUUUCGUACUCAAAAAGUAUGAU_ACUGGUGGUUUUACCAGCCAC__AAUGGCAACCACUACUCAUGCAAAACAAUAGCACGAAAAUGGUUUCUAUU_______________ ......(((((((.....)))))))...((((((....))))))......(((((.(((((.......(((.........)))......))))).)))))............... (-11.12 = -11.57 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:55 2006