
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,247,848 – 27,247,943 |
| Length | 95 |
| Max. P | 0.735716 |

| Location | 27,247,848 – 27,247,943 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.82 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -15.58 |
| Energy contribution | -14.95 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27247848 95 - 27905053 UUCGAAUAAAUUAAA--AGCGUGGGGUUAUGCGAAUAGCGAAUGCCCCGCAAGUGGAUUUUGAGUGCUCUGCAGGAACCUUUUGAGCUCAAGAUCUU---------- ...............--...(((((((..(((.....)))...)))))))....(((((((((..((((...((....))...))))))))))))).---------- ( -31.40) >DroSec_CAF1 20768 94 - 1 UUCGAAUAAAUUAAA--AGCAGGGGGGUAUGCGAAUAGCGAAUGCCCCGCAAGUGGAUUUUGAGUGCUCUGCAG-AACCUUUUGAGCUCAAGAUCCU---------- ...............--.((.((((.((.(((.....))).)).))))))....(((((((((((......(((-(....)))).))))))))))).---------- ( -31.30) >DroSim_CAF1 20899 93 - 1 UUCGAAUAAAUUAAA--AGCAUG-GGGUAUGCGAAUAGCGAAUGCCCCGCAAGUGGAUUUUGAGUGCUCUGCAG-AGCCCUUUGAGCUCAAGAUCCU---------- ...............--.((..(-((((((.((.....)).)))))))))....(((((((((((((((....)-))).......))))))))))).---------- ( -32.81) >DroEre_CAF1 20952 92 - 1 UUCGAAUAAAUUAAA--AGCUGG--GGUAUGCGAAUAGUGAAUGCCUCGCAAGUGGAUUUUGAGUACUCUGCAG-GACCUUUCGAGCUCAAGAUCUU---------- ...............--.((.((--(((((((.....))..)))))))))....((((((((((..(((.....-........))))))))))))).---------- ( -21.82) >DroYak_CAF1 21419 102 - 1 UUCGAAUAAAUUAAA--AGCCUG--GGUAUGCGAAUAGUGAAUGCCCCGCAAGUGGAUUUUGAGUGCUCUGCAG-AACCUUUCGAGCUCAAGAUCUUUCAAAUCAUC ...(((.........--.((..(--(((((((.....))..)))))).))....(((((((((..((((.....-........))))))))))))))))........ ( -22.52) >DroPer_CAF1 26466 86 - 1 UUGGUAAAAAUUCAAGAAACUUC-GGUCAUACGAAUGGUUGAUGCCUCGUCAUCGGAGAUUAAC-----------AACGGCUUGAGCUUGUAUUUCUC--------- ..............(((((((((-(((...((((..(((....))))))).)))))))....((-----------(..(((....)))))).))))).--------- ( -16.90) >consensus UUCGAAUAAAUUAAA__AGCAUG_GGGUAUGCGAAUAGCGAAUGCCCCGCAAGUGGAUUUUGAGUGCUCUGCAG_AACCUUUUGAGCUCAAGAUCCU__________ ..................((....((((((((.....))..)))))).))....(((((((((((....................)))))))))))........... (-15.58 = -14.95 + -0.63)

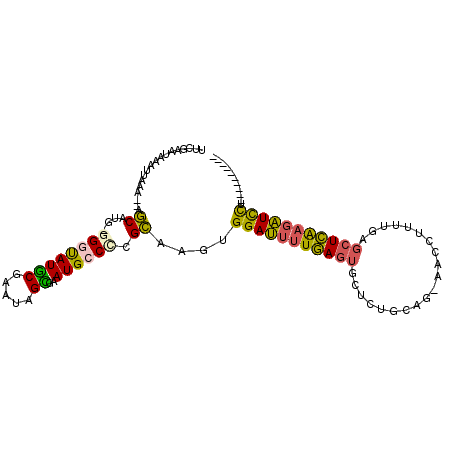

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:53 2006