| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,245,388 – 27,245,491 |
| Length | 103 |
| Max. P | 0.983879 |

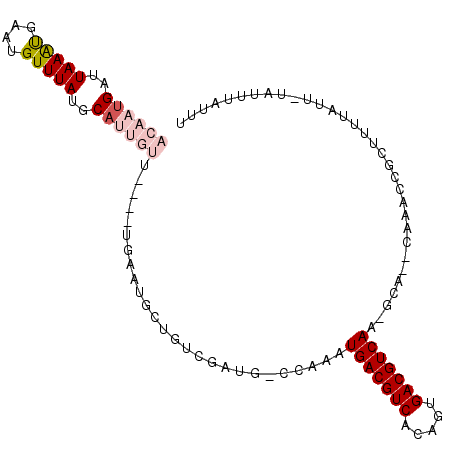
| Location | 27,245,388 – 27,245,491 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 72.51 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -12.68 |
| Energy contribution | -13.82 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27245388 103 + 27905053 GGAAUGAUUAAAUGAAUGUUUAUGCAGCGAUGUGGUGAAUGCUGUCGAUU-CGAAAUGACGUCACUGUGACGUCAA-GCA--CAAACCGCUUUUAUUUUAUUUAUUU .((((((..((((((((((....))).....(((((...((((.(((...-)))..(((((((.....))))))))-)))--...))))).)))))))...)))))) ( -27.30) >DroVir_CAF1 20415 87 + 1 ACAAUGAUUAAGUUAAUGUUUAUGCCUUGU------UAUUGCUGUCGAUGCCCAAAUGACGUCACAGUGACGUCAA-A-G--CGAUCCGCUUUUAUU---------- .(((((((...((..........))...))------)))))...............(((((((.....)))))))(-(-(--((...))))).....---------- ( -17.00) >DroPse_CAF1 22931 92 + 1 ACAAUGAUUAAACGAAUGUUUAUGCAUUGUU----UGU---------AUG-CCAAAUGACGUCACAGUGACGUCAAAGCAACGAAACCGCUUUUAUU-UAUUUAUUU .(((((..(((((....)))))..)))))((----(((---------.((-(....(((((((.....)))))))..))))))))............-......... ( -21.70) >DroSim_CAF1 18485 103 + 1 GGAAUGAUUAAAUGAAUGUUUAUGCAGCGAUGUGGUGAAUGCUGUCGAUU-CGAAAUGACGUCACUGUGACGUCAA-GCA--CAAACCGCUUUUAUUUUAUUUAUUU .((((((..((((((((((....))).....(((((...((((.(((...-)))..(((((((.....))))))))-)))--...))))).)))))))...)))))) ( -27.30) >DroWil_CAF1 13820 89 + 1 ACAAUGAUUAAAUGAAUGUUUAUGCAUUGU-GUUCUCUAGCUUGUCAAUU-CCAAAUGACGUCACUAUGACGUCAA-A-G--CAGACCGCUUUUU------------ ((((((..(((((....)))))..))))))-((..(((.((((.......-.....(((((((.....))))))))-)-)--))))..)).....------------ ( -20.90) >DroPer_CAF1 22914 92 + 1 ACAAUGAUUAAACGAAUGUUUAUGCAUUGUU----UGU---------AUG-CCAAAUGACGUCACAGUGACGUCAAAGCAACGAAACCGCUUUUAUU-UAUUUAUUU .(((((..(((((....)))))..)))))((----(((---------.((-(....(((((((.....)))))))..))))))))............-......... ( -21.70) >consensus ACAAUGAUUAAAUGAAUGUUUAUGCAUUGUU____UGAAUGCUGUCGAUG_CCAAAUGACGUCACAGUGACGUCAA_GCA__CAAACCGCUUUUAUU_UAUUUAUUU ((((((..(((((....)))))..))))))..........................(((((((.....)))))))................................ (-12.68 = -13.82 + 1.14)


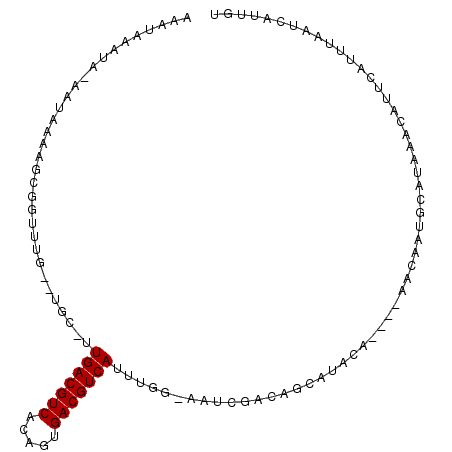
| Location | 27,245,388 – 27,245,491 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.51 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -10.20 |
| Energy contribution | -10.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27245388 103 - 27905053 AAAUAAAUAAAAUAAAAGCGGUUUG--UGC-UUGACGUCACAGUGACGUCAUUUCG-AAUCGACAGCAUUCACCACAUCGCUGCAUAAACAUUCAUUUAAUCAUUCC .................(((((.((--((.-.(((((((.....)))))))....(-((((....).))))..))))..)))))....................... ( -22.40) >DroVir_CAF1 20415 87 - 1 ----------AAUAAAAGCGGAUCG--C-U-UUGACGUCACUGUGACGUCAUUUGGGCAUCGACAGCAAUA------ACAAGGCAUAAACAUUAACUUAAUCAUUGU ----------.......((.(((.(--(-(-((((((((.....)))))))...)))))))....))....------.............................. ( -16.60) >DroPse_CAF1 22931 92 - 1 AAAUAAAUA-AAUAAAAGCGGUUUCGUUGCUUUGACGUCACUGUGACGUCAUUUGG-CAU---------ACA----AACAAUGCAUAAACAUUCGUUUAAUCAUUGU .........-...............((((((.(((((((.....)))))))...))-)).---------)).----.((((((..(((((....)))))..)))))) ( -23.00) >DroSim_CAF1 18485 103 - 1 AAAUAAAUAAAAUAAAAGCGGUUUG--UGC-UUGACGUCACAGUGACGUCAUUUCG-AAUCGACAGCAUUCACCACAUCGCUGCAUAAACAUUCAUUUAAUCAUUCC .................(((((.((--((.-.(((((((.....)))))))....(-((((....).))))..))))..)))))....................... ( -22.40) >DroWil_CAF1 13820 89 - 1 ------------AAAAAGCGGUCUG--C-U-UUGACGUCAUAGUGACGUCAUUUGG-AAUUGACAAGCUAGAGAAC-ACAAUGCAUAAACAUUCAUUUAAUCAUUGU ------------....(((.(((..--(-(-.(((((((.....)))))))...))-....)))..))).......-((((((..((((......))))..)))))) ( -20.50) >DroPer_CAF1 22914 92 - 1 AAAUAAAUA-AAUAAAAGCGGUUUCGUUGCUUUGACGUCACUGUGACGUCAUUUGG-CAU---------ACA----AACAAUGCAUAAACAUUCGUUUAAUCAUUGU .........-...............((((((.(((((((.....)))))))...))-)).---------)).----.((((((..(((((....)))))..)))))) ( -23.00) >consensus AAAUAAAUA_AAUAAAAGCGGUUUG__UGC_UUGACGUCACAGUGACGUCAUUUGG_AAUCGACAGCAUACA____AACAAUGCAUAAACAUUCAUUUAAUCAUUGU ................................(((((((.....)))))))........................................................ (-10.20 = -10.20 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:52 2006