
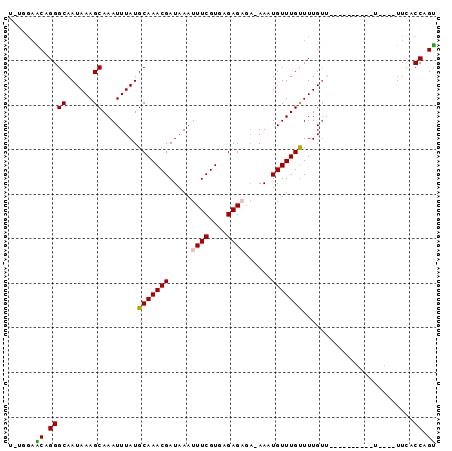
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,200,504 – 27,200,598 |
| Length | 94 |
| Max. P | 0.637692 |

| Location | 27,200,504 – 27,200,598 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -10.36 |
| Energy contribution | -12.08 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27200504 94 + 27905053 CAUGGAACCGGGCAAUAAAGCAAAUUUAUGCAAACGAUAAAUUUCGUGCGAGAGA-AAUUGUUUGUUUUGU-GUUUUGUGUUUUGUGUUCGCCAGU ..(((...((((((.(((((((....((((((((((((...((((....))))..-.)))))))))...))-).....)))))))))))))))).. ( -23.90) >DroVir_CAF1 20083 78 + 1 UUUGGAGCAGGGCAAUAAAGCAAAUUUAUGCAAACGAUAAAUUUCGUGAGAG--A-AAAUGUUUGUUUUGUU---------------UUCACCAGC .((((((((((((((....(((......)))..........((((....)))--)-......))))))))))---------------....)))). ( -16.90) >DroWil_CAF1 17795 76 + 1 ------ACAGGGCAAUAAAGCAAAUUUAUGCAAACGAUAAAUUUCGUGAGAGAGCGAAAUGUUUGUUUUGUU----------U----UUCACCAGU ------...((.....((((((((.....(((((((.....((((....))))......)))))))))))))----------)----)...))... ( -17.30) >DroYak_CAF1 14746 95 + 1 CAUGGAACCGGGCAAUAAAGCAAAUUUAUGCAAACGAUAAAUUUCGUGCGAGAGA-AAUUGUUUGCUUUGUUUGGUUUUGUUUUGGGUUCGCCAGU ....(((((.((.((((((((((((....(((((((((...((((....))))..-.)))))))))...)))).)))))))))).)))))...... ( -28.90) >DroMoj_CAF1 17843 78 + 1 UUAGGAGCAGGGCAAUAAAGCAAAUUUAUGCAAACGAUAAAUUUCGUGAGAG--A-AAAUGUUUGUUUUGUU---------------UUCACCAGC ...((((((((((((....(((......)))..........((((....)))--)-......))))))))))---------------))....... ( -16.50) >DroAna_CAF1 15712 84 + 1 U-CCGAGCCGGGCAAUAAAGCAAAUUUAUGCAAACGAUAAAUUUCGUGAGAGAGA-AAAUGUUUGUUUUG----------UUUUGUUUUUGCCAGU .-........(((((..(((((((.....(((((((.....((((....))))..-...)))))))....----------.))))))))))))... ( -24.10) >consensus U_UGGAACAGGGCAAUAAAGCAAAUUUAUGCAAACGAUAAAUUUCGUGAGAGAGA_AAAUGUUUGUUUUGUU__________U____UUCACCAGU ......((.((((......))........(((((((.....((((....))))......))))))).........................)).)) (-10.36 = -12.08 + 1.72)


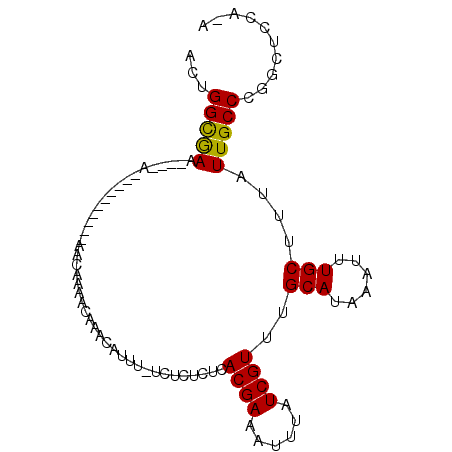
| Location | 27,200,504 – 27,200,598 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -12.17 |
| Consensus MFE | -9.54 |
| Energy contribution | -9.15 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27200504 94 - 27905053 ACUGGCGAACACAAAACACAAAAC-ACAAAACAAACAAUU-UCUCUCGCACGAAAUUUAUCGUUUGCAUAAAUUUGCUUUAUUGCCCGGUUCCAUG ((((((((................-...((((....((((-((........))))))....))))(((......)))....))).)))))...... ( -12.80) >DroVir_CAF1 20083 78 - 1 GCUGGUGAA---------------AACAAAACAAACAUUU-U--CUCUCACGAAAUUUAUCGUUUGCAUAAAUUUGCUUUAUUGCCCUGCUCCAAA ((.((..(.---------------....((((......((-(--(......))))......))))(((......)))....)..))..))...... ( -10.90) >DroWil_CAF1 17795 76 - 1 ACUGGUGAA----A----------AACAAAACAAACAUUUCGCUCUCUCACGAAAUUUAUCGUUUGCAUAAAUUUGCUUUAUUGCCCUGU------ ...((..(.----.----------....((((....((((((........)))))).....))))(((......)))....)..))....------ ( -10.50) >DroYak_CAF1 14746 95 - 1 ACUGGCGAACCCAAAACAAAACCAAACAAAGCAAACAAUU-UCUCUCGCACGAAAUUUAUCGUUUGCAUAAAUUUGCUUUAUUGCCCGGUUCCAUG ....(.(((((...................((((((((((-((........))))))....))))))........((......))..))))))... ( -16.60) >DroMoj_CAF1 17843 78 - 1 GCUGGUGAA---------------AACAAAACAAACAUUU-U--CUCUCACGAAAUUUAUCGUUUGCAUAAAUUUGCUUUAUUGCCCUGCUCCUAA ((.((..(.---------------....((((......((-(--(......))))......))))(((......)))....)..))..))...... ( -10.90) >DroAna_CAF1 15712 84 - 1 ACUGGCAAAAACAAAA----------CAAAACAAACAUUU-UCUCUCUCACGAAAUUUAUCGUUUGCAUAAAUUUGCUUUAUUGCCCGGCUCGG-A .(.(((((........----------..............-........((((......))))..(((......)))....))))).)......-. ( -11.30) >consensus ACUGGCGAA____A__________AACAAAACAAACAUUU_UCUCUCUCACGAAAUUUAUCGUUUGCAUAAAUUUGCUUUAUUGCCCGGCUCCA_A ...(((((.........................................((((......))))..(((......)))....))))).......... ( -9.54 = -9.15 + -0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:42 2006