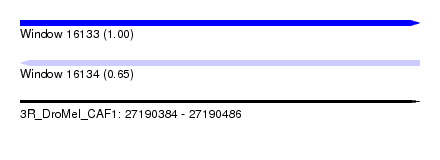
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,190,384 – 27,190,486 |
| Length | 102 |
| Max. P | 0.996641 |

| Location | 27,190,384 – 27,190,486 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.23 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.57 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27190384 102 + 27905053 AUGGGAAUAAAA------AUAUGAAAUUGCCAAUUGAUUUAUUGGCAGAAAACAUUUUUCCAUGUCAAAUAGUGCGGAAAAUGUACAAAGUG-AGAAACAAA-GGGAAAC (((((((.....------..(((...((((((((......))))))))....))).))))))).(((....(((((.....)))))....))-)........-....... ( -19.30) >DroSec_CAF1 3978 102 + 1 AUGGGCAUAAAA------AUAUGAAAUUGCCAAUUGAUUUAUUGGCAGAAAACAUUUUUCCAUGUCAAAUAGUGCGGAAAAUGUACAAAGUG-AGAAACAAA-GGGAAAC ...(((((..((------(.(((...((((((((......))))))))....))).)))..))))).....(((((.....)))))......-.........-....... ( -19.90) >DroSim_CAF1 3739 102 + 1 AUGGGCAUAAAA------AUAUGAAAUUGCCAAUUGAUUUAUUGGCAGAAAACAUUUUUCCAUGUCAAAUAGUGCGGAAAAUGUACAAAGUG-AGAAACAAA-GGGAAAC ...(((((..((------(.(((...((((((((......))))))))....))).)))..))))).....(((((.....)))))......-.........-....... ( -19.90) >DroEre_CAF1 5575 102 + 1 AUGGUCAUAAAA------AUAUGAAAUUGCCAAUUGAUUUAUUGGCAGAAAACAUUUUUCCAUGUCAAAUAGUGCGGAAAACGUACAAAGUG-AGAAACAAA-GGGAAAC ....(((((...------.)))))..((((((((......)))))))).......((((((..........(((((.....)))))...((.-....))...-)))))). ( -23.70) >DroYak_CAF1 4137 108 + 1 AGGGCCAUAAAAAUAGAAAUAUGAAAUUGCCAAUUGAUUUAUUGGCAGAAAACAUUUUUCCAUGUCAAAUAGUGCGGAAAAUGUACAAAGUG-AGUAACAAA-GGGAAAC ....((......(((....)))....((((((((......))))))))...((((((((((((........))).)))))))))........-.........-))..... ( -21.20) >DroAna_CAF1 3924 104 + 1 AAGAGGUAAAAA------AUAUGAAAUUGCCAAUUGAUUUAUUGGCAGAAAACAUUUUUCCAUGUCAAAUAGUGCGGAAAAUGUAAAAAAAGCUGAAACAAAAGAAAAAC ....(((.....------........((((((((......))))))))...((((((((((((........))).))))))))).......)))................ ( -20.10) >consensus AUGGGCAUAAAA______AUAUGAAAUUGCCAAUUGAUUUAUUGGCAGAAAACAUUUUUCCAUGUCAAAUAGUGCGGAAAAUGUACAAAGUG_AGAAACAAA_GGGAAAC ..........................((((((((......))))))))...((((((((((((........))).))))))))).......................... (-18.40 = -18.57 + 0.17)


| Location | 27,190,384 – 27,190,486 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.23 |
| Mean single sequence MFE | -15.59 |
| Consensus MFE | -13.07 |
| Energy contribution | -12.93 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27190384 102 - 27905053 GUUUCCC-UUUGUUUCU-CACUUUGUACAUUUUCCGCACUAUUUGACAUGGAAAAAUGUUUUCUGCCAAUAAAUCAAUUGGCAAUUUCAUAU------UUUUAUUCCCAU .(((((.-..((((...-.....(((.........)))......)))).))))).(((.....(((((((......)))))))....)))..------............ ( -13.64) >DroSec_CAF1 3978 102 - 1 GUUUCCC-UUUGUUUCU-CACUUUGUACAUUUUCCGCACUAUUUGACAUGGAAAAAUGUUUUCUGCCAAUAAAUCAAUUGGCAAUUUCAUAU------UUUUAUGCCCAU .......-.........-............................((((((((.(((.....(((((((......)))))))....))).)------)))))))..... ( -15.30) >DroSim_CAF1 3739 102 - 1 GUUUCCC-UUUGUUUCU-CACUUUGUACAUUUUCCGCACUAUUUGACAUGGAAAAAUGUUUUCUGCCAAUAAAUCAAUUGGCAAUUUCAUAU------UUUUAUGCCCAU .......-.........-............................((((((((.(((.....(((((((......)))))))....))).)------)))))))..... ( -15.30) >DroEre_CAF1 5575 102 - 1 GUUUCCC-UUUGUUUCU-CACUUUGUACGUUUUCCGCACUAUUUGACAUGGAAAAAUGUUUUCUGCCAAUAAAUCAAUUGGCAAUUUCAUAU------UUUUAUGACCAU .......-........(-((....((.((.....)).))....)))((((((((.(((.....(((((((......)))))))....))).)------)))))))..... ( -17.20) >DroYak_CAF1 4137 108 - 1 GUUUCCC-UUUGUUACU-CACUUUGUACAUUUUCCGCACUAUUUGACAUGGAAAAAUGUUUUCUGCCAAUAAAUCAAUUGGCAAUUUCAUAUUUCUAUUUUUAUGGCCCU .......-..(((.((.-......))))).........((((..((.(((((((.(((.....(((((((......)))))))....))).)))))))))..)))).... ( -17.80) >DroAna_CAF1 3924 104 - 1 GUUUUUCUUUUGUUUCAGCUUUUUUUACAUUUUCCGCACUAUUUGACAUGGAAAAAUGUUUUCUGCCAAUAAAUCAAUUGGCAAUUUCAUAU------UUUUUACCUCUU ((((((((..((((...((................)).......)))).))))))))......(((((((......))))))).........------............ ( -14.29) >consensus GUUUCCC_UUUGUUUCU_CACUUUGUACAUUUUCCGCACUAUUUGACAUGGAAAAAUGUUUUCUGCCAAUAAAUCAAUUGGCAAUUUCAUAU______UUUUAUGCCCAU ..........................((((((((((............))).)))))))....(((((((......)))))))........................... (-13.07 = -12.93 + -0.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:40 2006