
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
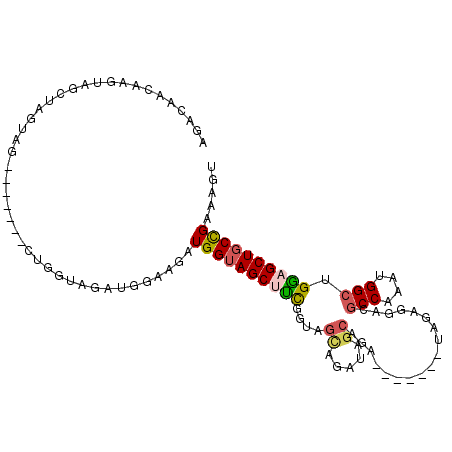
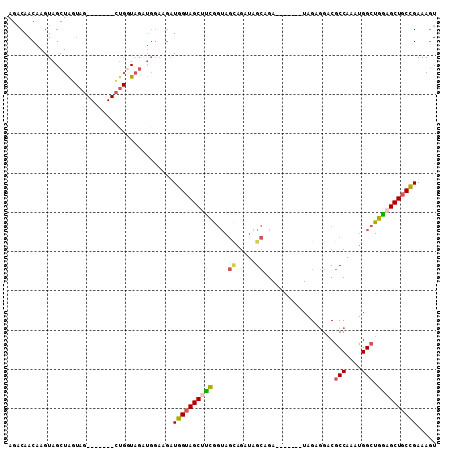
| Location | 27,180,927 – 27,181,022 |
| Length | 95 |
| Max. P | 0.828005 |

| Location | 27,180,927 – 27,181,022 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -25.49 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.45 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.828005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
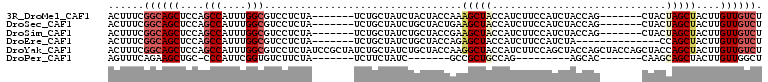
>3R_DroMel_CAF1 27180927 95 + 27905053 AGACAACAAGUAGCUAGUAG-------CUGGUAGAUGGAAGAUGGUAGCUUUGGUAGUAGAUAGCAGA-------UAGAGGACGCCAAAUGGCUGGAGCUGCCGAAAGU ...((.(.(.((((.....)-------))).).).)).....(((((((((..((........((...-------........))......))..)))))))))..... ( -23.24) >DroSec_CAF1 77284 95 + 1 AGACAACAAGUAGCUAGUAG-------CUGGUAGAUGGAAGAUGGUAGCUUCAGUAGCAGAUAGCAGA-------UAGAGGACGCCAAAUGGCUGGAGCUGCCGAAAGU ...((.(.(.((((.....)-------))).).).)).....(((((((((((...((.....))...-------........(((....))))))))))))))..... ( -27.40) >DroSim_CAF1 80608 95 + 1 AGACAACAAGUAGCUAGUAG-------CUGGUAGAUGGAAGAUGGUAGCUUCGGUAGCAGAUAGCAGA-------UAGAGGACGCCAAAUGGCUGGAGCUGCCGAAAGU ...((.(.(.((((.....)-------))).).).)).....(((((((((((...((.....))...-------........(((....))))))))))))))..... ( -26.70) >DroEre_CAF1 78232 88 + 1 AGACAACAAGUAGCUGG--------------UAGAUGGAAGAUGGUAGCUCUGGUAGCAGAUAGCAGA-------UAGAGGACGCCAAAUGGCUGGAGCUGCCGAAAGU .................--------------...........((((((((((....((.....))...-------........(((....))).))))))))))..... ( -22.30) >DroYak_CAF1 81572 109 + 1 AGACAACAAGUAGCUGGUAGCUGGUAGCUGGUAGCUGGAAGAUGGUAGCCUUGGUAGCAGAUAGCAGAUAGCGGAUAGAGGACGCCAAAUGGCUGGAGCUGCCGAAAGU ..........((((((.((((.....)))).)))))).............((((((((...((((.....(((.........)))......))))..)))))))).... ( -32.90) >DroPer_CAF1 150262 78 + 1 AGCCAACAAGUAGCUGCUUG-------GUGCU---------CUGGCAGCGGC-------GAUAGAAGA-------UAGAAGACACCGAAUGGG-GCAGCUUCUGAAACU .(((..(((((....)))))-------.(((.---------...)))..)))-------.........-------((((((...((.....))-....))))))..... ( -20.40) >consensus AGACAACAAGUAGCUAGUAG_______CUGGUAGAUGGAAGAUGGUAGCUUCGGUAGCAGAUAGCAGA_______UAGAGGACGCCAAAUGGCUGGAGCUGCCGAAAGU ..........................................((((((((((....((.....))..................(((....))).))))))))))..... (-16.34 = -16.45 + 0.11)

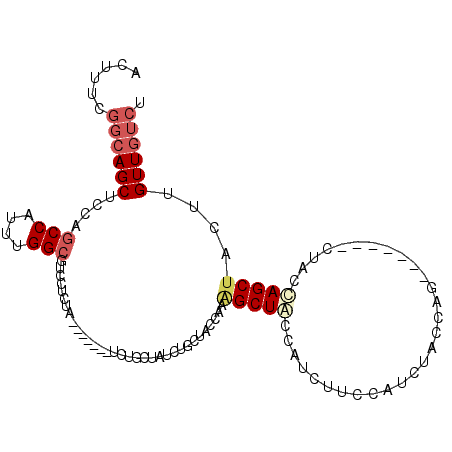
| Location | 27,180,927 – 27,181,022 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -18.60 |
| Consensus MFE | -10.29 |
| Energy contribution | -11.18 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27180927 95 - 27905053 ACUUUCGGCAGCUCCAGCCAUUUGGCGUCCUCUA-------UCUGCUAUCUACUACCAAAGCUACCAUCUUCCAUCUACCAG-------CUACUAGCUACUUGUUGUCU ......((((((....(((....)))........-------..................(((((..................-------....)))))....)))))). ( -14.35) >DroSec_CAF1 77284 95 - 1 ACUUUCGGCAGCUCCAGCCAUUUGGCGUCCUCUA-------UCUGCUAUCUGCUACUGAAGCUACCAUCUUCCAUCUACCAG-------CUACUAGCUACUUGUUGUCU ......((((((....(((....)))........-------...((((...(((...((((.......))))........))-------)...)))).....)))))). ( -17.80) >DroSim_CAF1 80608 95 - 1 ACUUUCGGCAGCUCCAGCCAUUUGGCGUCCUCUA-------UCUGCUAUCUGCUACCGAAGCUACCAUCUUCCAUCUACCAG-------CUACUAGCUACUUGUUGUCU ......((((((....(((....)))........-------...((((...(((...((((.......))))........))-------)...)))).....)))))). ( -17.80) >DroEre_CAF1 78232 88 - 1 ACUUUCGGCAGCUCCAGCCAUUUGGCGUCCUCUA-------UCUGCUAUCUGCUACCAGAGCUACCAUCUUCCAUCUA--------------CCAGCUACUUGUUGUCU ......((.(((((..(((....)))........-------...((.....)).....))))).))............--------------.((((.....))))... ( -18.90) >DroYak_CAF1 81572 109 - 1 ACUUUCGGCAGCUCCAGCCAUUUGGCGUCCUCUAUCCGCUAUCUGCUAUCUGCUACCAAGGCUACCAUCUUCCAGCUACCAGCUACCAGCUACCAGCUACUUGUUGUCU ......((((((...((((...(((((.........)))))...((.....))......))))..........((((...(((.....)))...))))....)))))). ( -25.00) >DroPer_CAF1 150262 78 - 1 AGUUUCAGAAGCUGC-CCCAUUCGGUGUCUUCUA-------UCUUCUAUC-------GCCGCUGCCAG---------AGCAC-------CAAGCAGCUACUUGUUGGCU ((((.(((.((((((-.......((((.......-------........)-------)))(((.....---------)))..-------...)))))).).))..)))) ( -17.76) >consensus ACUUUCGGCAGCUCCAGCCAUUUGGCGUCCUCUA_______UCUGCUAUCUGCUACCAAAGCUACCAUCUUCCAUCUACCAG_______CUACCAGCUACUUGUUGUCU ......((((((....(((....))).................................(((((.............................)))))....)))))). (-10.29 = -11.18 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:36 2006