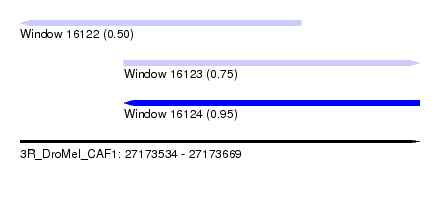
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,173,534 – 27,173,669 |
| Length | 135 |
| Max. P | 0.950086 |

| Location | 27,173,534 – 27,173,629 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.54 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.70 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27173534 95 - 27905053 GGGGCAA-AUAAAAACAAAGCUGGCUGAAAUGGAAAAUAAGAAGUGAGUCCGAGUCGCCGGCAAUGGGGGUCAGGUCAGAUGCUGGAAAUGAUGCU----- ..((((.-...........((((((.((..((((..((.....))...))))..))))))))..(.((.(((......))).)).)......))))----- ( -24.40) >DroSec_CAF1 70062 96 - 1 GGGGCAAAAUAAAAACAAAGCUGGCUGAAAUGGAAAAUAAGAAGCGAGUCCGAGUCGCCGCCAAUGGGGGUCAGGUCAGAUGCUGGAAAUGAUGCU----- ..((((............(((...((((..((........((..((....))..))(((.((...)).)))))..))))..)))........))))----- ( -20.25) >DroSim_CAF1 72942 96 - 1 GGGGCAAAAUAAAAACAAAGCCCGCUGAAAUGGAAAAUAAGAAGCGAGUCCGAGUCGCCGGCAAUGGGGGUCAGGUCAGAUGCUGGAAAUGAUGCU----- .((((..............))))((((...(....).......((((.......))))))))....((.((((..((((...))))...)))).))----- ( -20.84) >DroEre_CAF1 70832 100 - 1 GGAGCAA-AUAAAAACAAAGCUGGCUGAAAUGGAAAAUAAGAAGUGAGUCCGAGUCGUCGGCAAUGGGGGUCAGGUCAGAUGCUGGAGCUGAUGCUCGACU .((((..-...........((((((.((..((((..((.....))...))))..)))))))).......(((((.((........)).))))))))).... ( -28.70) >consensus GGGGCAA_AUAAAAACAAAGCUGGCUGAAAUGGAAAAUAAGAAGCGAGUCCGAGUCGCCGGCAAUGGGGGUCAGGUCAGAUGCUGGAAAUGAUGCU_____ ..((((.............((((((.((..((((..((.....))...))))..))))))))..(.((.(((......))).)).)......))))..... (-20.26 = -20.70 + 0.44)

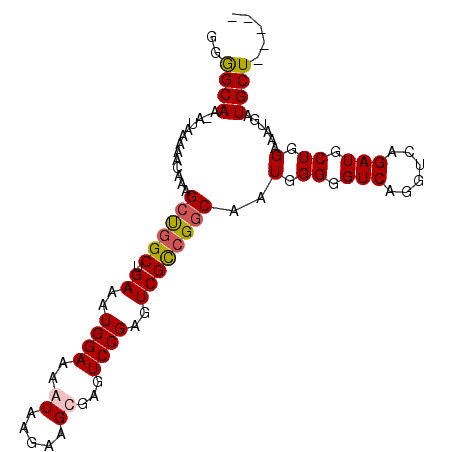
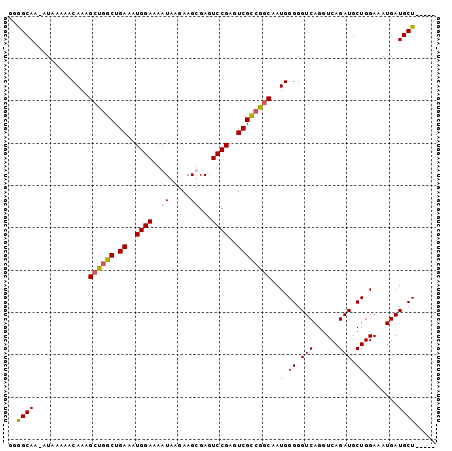
| Location | 27,173,569 – 27,173,669 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.05 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -17.82 |
| Energy contribution | -19.22 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27173569 100 + 27905053 CCGGCGACUCGGACUCACUUCUUAUUUU-------CCAUUUCAGCCAGCUUUGUUUUUAU-UUGCCCCUCCAAGGGCAAUAAGCUGGCAUUUCUUGUUAGCCGCCUUG .(((((((..(((..............)-------))......((((((((.........-((((((......)))))).)))))))).......))).))))..... ( -30.84) >DroSec_CAF1 70097 101 + 1 GCGGCGACUCGGACUCGCUUCUUAUUUU-------CCAUUUCAGCCAGCUUUGUUUUUAUUUUGCCCCUCCAAGGGCAAUAAGCUGGCGUUUCUUGUUAGCCGCCUUG ((((((((..(((..............)-------))......((((((((..........((((((......)))))).)))))))).......))).))))).... ( -34.24) >DroSim_CAF1 72977 101 + 1 CCGGCGACUCGGACUCGCUUCUUAUUUU-------CCAUUUCAGCGGGCUUUGUUUUUAUUUUGCCCCUCCAAGGGCAAUAAGCUGGCGUUUCUUGUUAGCCGCCUUG .(((((((..(((..((((.((((((.(-------((.((..((.((((...((....))...))))))..))))).))))))..))))..))).))).))))..... ( -30.60) >DroEre_CAF1 70872 100 + 1 CCGACGACUCGGACUCACUUCUUAUUUU-------CCAUUUCAGCCAGCUUUGUUUUUAU-UUGCUCCUGCAAGGGCAAUAAGCUGGCGUUUCUUGUUAGCCGCCUUG ((((....))))................-------........((((((((((((....(-((((....)))))))))..)))))))).................... ( -23.10) >DroYak_CAF1 73766 94 + 1 -------------CUCACUUCUUAUUUUCCAUUUUCCAUUUCAGGCAGCUUUGUUUUUAC-UUACUCCUGCAAGGGCAAUAAGCUGGCGUUUCUUGUUAGCCGCCUUG -------------.............................((((((((((((((((.(-........).)))))))..)))))(((...........))))))).. ( -17.70) >consensus CCGGCGACUCGGACUCACUUCUUAUUUU_______CCAUUUCAGCCAGCUUUGUUUUUAU_UUGCCCCUCCAAGGGCAAUAAGCUGGCGUUUCUUGUUAGCCGCCUUG ..((((.....................................((((((((..........((((((......)))))).))))))))(((.......)))))))... (-17.82 = -19.22 + 1.40)

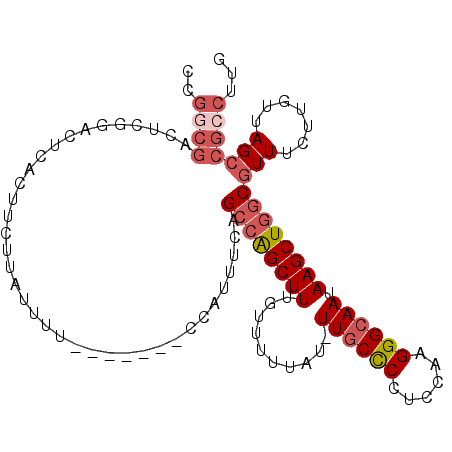
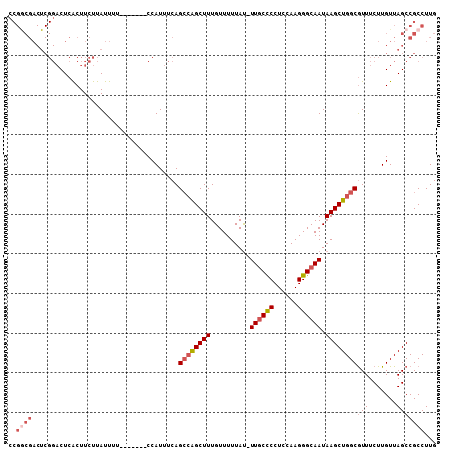
| Location | 27,173,569 – 27,173,669 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.05 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -20.58 |
| Energy contribution | -22.94 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
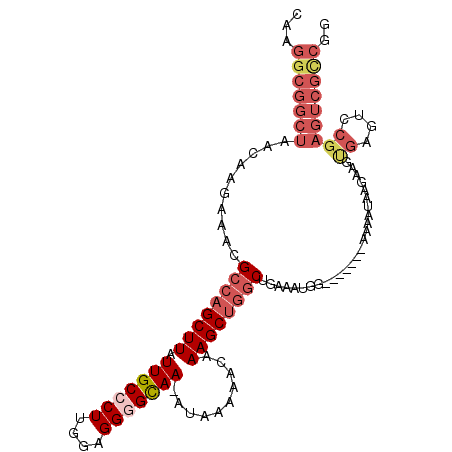
>3R_DroMel_CAF1 27173569 100 - 27905053 CAAGGCGGCUAACAAGAAAUGCCAGCUUAUUGCCCUUGGAGGGGCAA-AUAAAAACAAAGCUGGCUGAAAUGG-------AAAAUAAGAAGUGAGUCCGAGUCGCCGG ...(((((((..........((((((((.(((((((....)))))))-.........)))))))).....(((-------(..((.....))...))))))))))).. ( -37.30) >DroSec_CAF1 70097 101 - 1 CAAGGCGGCUAACAAGAAACGCCAGCUUAUUGCCCUUGGAGGGGCAAAAUAAAAACAAAGCUGGCUGAAAUGG-------AAAAUAAGAAGCGAGUCCGAGUCGCCGC ...(((((((..........((((((((.(((((((....)))))))..........)))))))).....(((-------(..............))))))))))).. ( -36.14) >DroSim_CAF1 72977 101 - 1 CAAGGCGGCUAACAAGAAACGCCAGCUUAUUGCCCUUGGAGGGGCAAAAUAAAAACAAAGCCCGCUGAAAUGG-------AAAAUAAGAAGCGAGUCCGAGUCGCCGG ...(((((((.....((..(((...((((((..((((..((((((..............)))).))..)).))-------..))))))..)))..))..))))))).. ( -33.04) >DroEre_CAF1 70872 100 - 1 CAAGGCGGCUAACAAGAAACGCCAGCUUAUUGCCCUUGCAGGAGCAA-AUAAAAACAAAGCUGGCUGAAAUGG-------AAAAUAAGAAGUGAGUCCGAGUCGUCGG ...(((((((..........((((((((.((((((.....)).))))-.........)))))))).....(((-------(..((.....))...))))))))))).. ( -30.10) >DroYak_CAF1 73766 94 - 1 CAAGGCGGCUAACAAGAAACGCCAGCUUAUUGCCCUUGCAGGAGUAA-GUAAAAACAAAGCUGCCUGAAAUGGAAAAUGGAAAAUAAGAAGUGAG------------- ..((((((((.....(......).(((((((.((......)))))))-))........)))))))).............................------------- ( -19.00) >consensus CAAGGCGGCUAACAAGAAACGCCAGCUUAUUGCCCUUGGAGGGGCAA_AUAAAAACAAAGCUGGCUGAAAUGG_______AAAAUAAGAAGUGAGUCCGAGUCGCCGG ...(((((((..........((((((((.(((((((....)))))))..........))))))))..........................((....))))))))).. (-20.58 = -22.94 + 2.36)

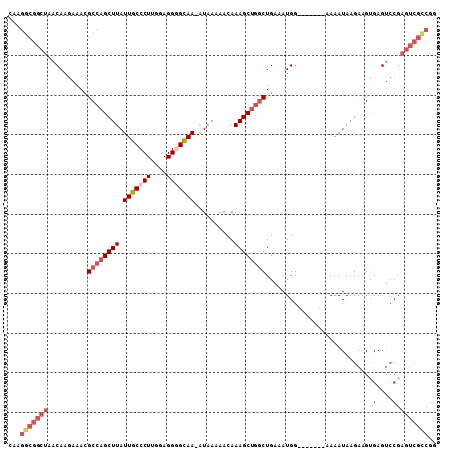
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:31 2006