
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,161,718 – 27,161,808 |
| Length | 90 |
| Max. P | 0.844511 |

| Location | 27,161,718 – 27,161,808 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -19.10 |
| Energy contribution | -20.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
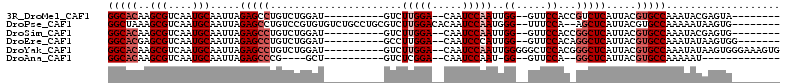
>3R_DroMel_CAF1 27161718 90 + 27905053 GGCACAAGCGUCAAUGCAAUUAGAGCCUGUCUGGAU----------GUCUUGGA--CAAUCCAAUUGG--GUUCCACCGUCUCAUUACGUGCCAAAUACGAGUA-------- (((((..(((....))).....(((((((..(((((----------(((...))--).)))))..)))--))))..............)))))...........-------- ( -27.70) >DroPse_CAF1 75044 100 + 1 GGCUAAAGCGUCAAUGCAAUUAGAGCCUGUCCGUGUGUCUGCCUGCGUCUUGGACACAAUCCAAUGGG--UUUCCA--AGCUCAUUACGUGCCAAAAAUAAGUG-------- (((....((((.((((...((.((((((((...(((((((...........))))))).....)))))--)))..)--)...)))))))))))...........-------- ( -23.60) >DroSim_CAF1 61116 90 + 1 GGCACAAGCGUCAAUGCAAUUAGAGCCUGUCUGGAU----------GUCUUGGA--CAAUCCAAUUGG--GUUCCACCGGCUCAUUACGUGCCAAAUACGAGUG-------- (((((..(((....))).....(((((....(((((----------(((...))--).)))))...((--......))))))).....)))))...........-------- ( -30.00) >DroEre_CAF1 59162 91 + 1 GGCACGAGCGUCAAUGCAAUUAGAGCCUGUCUGGAU----------GCCUUGGA--CAAUCCCAUUGG--GUUCCACAGGCUCAUUACGUGCCAAAUAUAAGUGG------- ((((((.(((....))).....((((((((..(((.----------(((((((.--.....)))..))--)))))))))))))....))))))............------- ( -37.80) >DroYak_CAF1 61518 100 + 1 GGCACAAGCGUCAAUGCAAUUAGAGCCUGUCUGGAU----------GUCUUGGA--CAAUCCAAUUGGGGGCUCCACGGGCUCAUUACGUGCCAAAUAUAAGUGGGAAAGUG (((((..(((....))).....((((((((..(((.----------(((((((.--....))).....))))))))))))))).....)))))................... ( -34.00) >DroAna_CAF1 63862 78 + 1 GGCACAAGCGUCAAUGCAAUUAGAGCCCG----GCU----------GUCUCGGA--CAAUCCAAU-GG--GUUCCA--GGCUCAUUACGUGCCAAAAAU------------- (((((..(((....))).....(((((.(----(..----------..(((((.--....))...-))--)..)).--))))).....)))))......------------- ( -25.40) >consensus GGCACAAGCGUCAAUGCAAUUAGAGCCUGUCUGGAU__________GUCUUGGA__CAAUCCAAUUGG__GUUCCAC_GGCUCAUUACGUGCCAAAUAUAAGUG________ (((((..(((....))).....(((((......................(((((.....)))))..((.....))...))))).....)))))................... (-19.10 = -20.10 + 1.00)
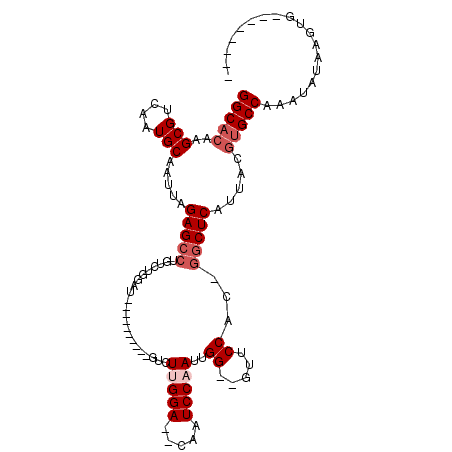

| Location | 27,161,718 – 27,161,808 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27161718 90 - 27905053 --------UACUCGUAUUUGGCACGUAAUGAGACGGUGGAAC--CCAAUUGGAUUG--UCCAAGAC----------AUCCAGACAGGCUCUAAUUGCAUUGACGCUUGUGCC --------...........(((((((((((......((((.(--(...((((((.(--((...)))----------))))))...)).))))....))))).....)))))) ( -22.20) >DroPse_CAF1 75044 100 - 1 --------CACUUAUUUUUGGCACGUAAUGAGCU--UGGAAA--CCCAUUGGAUUGUGUCCAAGACGCAGGCAGACACACGGACAGGCUCUAAUUGCAUUGACGCUUUAGCC --------...........(((.......(((((--((....--((...((..(((((((...))))))).....))...)).))))))).....((......))....))) ( -22.40) >DroSim_CAF1 61116 90 - 1 --------CACUCGUAUUUGGCACGUAAUGAGCCGGUGGAAC--CCAAUUGGAUUG--UCCAAGAC----------AUCCAGACAGGCUCUAAUUGCAUUGACGCUUGUGCC --------...........((((((....(((((((......--))..((((((.(--((...)))----------))))))...))))).....((......)).)))))) ( -26.70) >DroEre_CAF1 59162 91 - 1 -------CCACUUAUAUUUGGCACGUAAUGAGCCUGUGGAAC--CCAAUGGGAUUG--UCCAAGGC----------AUCCAGACAGGCUCUAAUUGCAUUGACGCUCGUGCC -------............((((((....(((((((((((.(--((...)))..((--(.....))----------))))..)))))))).....((......)).)))))) ( -30.90) >DroYak_CAF1 61518 100 - 1 CACUUUCCCACUUAUAUUUGGCACGUAAUGAGCCCGUGGAGCCCCCAAUUGGAUUG--UCCAAGAC----------AUCCAGACAGGCUCUAAUUGCAUUGACGCUUGUGCC ...................(((((((((((......(((((((.....((((((.(--((...)))----------))))))...)))))))....))))).....)))))) ( -27.50) >DroAna_CAF1 63862 78 - 1 -------------AUUUUUGGCACGUAAUGAGCC--UGGAAC--CC-AUUGGAUUG--UCCGAGAC----------AGC----CGGGCUCUAAUUGCAUUGACGCUUGUGCC -------------......((((((....(((((--(((...--..-.((((....--.))))...----------..)----))))))).....((......)).)))))) ( -28.90) >consensus ________CACUUAUAUUUGGCACGUAAUGAGCC_GUGGAAC__CCAAUUGGAUUG__UCCAAGAC__________AUCCAGACAGGCUCUAAUUGCAUUGACGCUUGUGCC ...................((((((....(((((...((.....))..(((((.....)))))......................))))).....((......)).)))))) (-18.34 = -18.62 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:28 2006