
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,154,466 – 27,154,571 |
| Length | 105 |
| Max. P | 0.592639 |

| Location | 27,154,466 – 27,154,571 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.97 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -19.99 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27154466 105 - 27905053 A----UAUAUGCGC-----------ACAGGAAGAAUGUCGUAAAAAAGUUUUACGAUCAAACAUUGCACUUUAUGUGCUUUUAUGUAAAAUUACCGUACAACAUUGGCAACAUUGUCAGC .----.......((-----------...((......((((((((.....))))))))...((((.((((.....))))....)))).......)).....(((.((....)).)))..)) ( -21.00) >DroPse_CAF1 72893 116 - 1 A----UAUAUGCGCGCUGCUGUGCCACAGGAAAAAUGUCGUAAAAAAGUUUUACGAUCAAACAUUGCACUUUAUGUGCUUUUAUGUAAAAUUACCGUACAACAUUGGCAACAUUGUCUGC .----....(((((((....))))....((......((((((((.....))))))))...((((.((((.....))))....)))).......)))))(((((.((....)).))).)). ( -26.20) >DroGri_CAF1 82947 109 - 1 AUAUAUAUAUGCGU-----------ACAGGAAAAAUGUCGUAAAAAAGUUUUACGAUCAAACAUUGCACUUUAUGUGCUUUUAUGUAAAAUUACCGUACAACAUUGGCAACAUUGUCAGC ..........((((-----------((.((......((((((((.....))))))))...((((.((((.....))))....)))).......)))))).(((.((....)).)))..)) ( -25.10) >DroWil_CAF1 72414 105 - 1 A----UAUAUGCGC-----------ACAGGAAAAAUGUCGUAAAAAAGUUUUACGAUCAAACAUUGCACUUUAUGUGCUUUUAUGUAAAAUUACCGUACAGCAUUGGCAACAUUGUCAGC .----...((((..-----------...((......((((((((.....))))))))...((((.((((.....))))....)))).......)).....))))(((((....))))).. ( -23.20) >DroMoj_CAF1 64204 105 - 1 A----UAUAUGCGU-----------ACAGGAAAAAUGUCGUAAAAAAGUUUUACGAUCAAACAUUGCAGUUUAUGUGCUUUUAUGUAAAAUUACCGUACAACAUUGGCAACAUUGUCAGC .----.....((((-----------((.((......((((((((.....))))))))...((((...(((......)))...)))).......)))))).(((.((....)).)))..)) ( -20.90) >DroPer_CAF1 129013 116 - 1 A----UAUAUGCGCGCUGCUGUGCCACAGGAAAAAUGUCGUAAAAAAGUUUUACGAUCAAACAUUGCACUUUAUGUGCUUUUAUGUAAAAUUACCGUACAACAUUGGCAACAUUGUCAGC .----.........((((.(((((....((......((((((((.....))))))))...((((.((((.....))))....)))).......)))))))(((.((....)).))))))) ( -26.50) >consensus A____UAUAUGCGC___________ACAGGAAAAAUGUCGUAAAAAAGUUUUACGAUCAAACAUUGCACUUUAUGUGCUUUUAUGUAAAAUUACCGUACAACAUUGGCAACAUUGUCAGC ..........((................((......((((((((.....))))))))...((((.((((.....))))....)))).......)).....(((.((....)).)))..)) (-19.99 = -20.02 + 0.03)

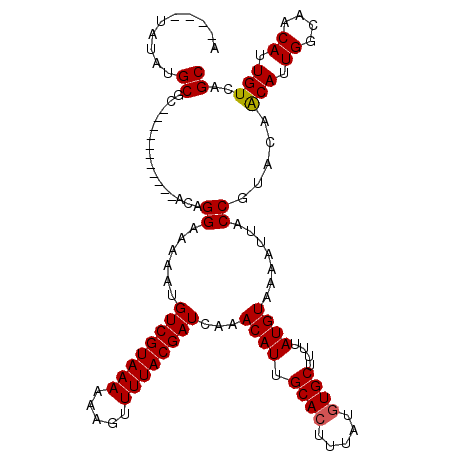

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:26 2006