| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,150,644 – 27,150,734 |
| Length | 90 |
| Max. P | 0.665637 |

| Location | 27,150,644 – 27,150,734 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
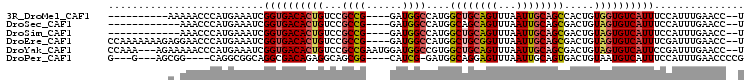
>3R_DroMel_CAF1 27150644 90 + 27905053 ----------AAAAACCCAUGAAAUCGGUGACACUGUCCGCCG----GAUGGCCAUGGCUGCAGUUUAAUUGCAGCCACUGUGGUGUCAUUUCCAUUUGAACC--U ----------................((((((((..(..(((.----...)))..((((((((((...))))))))))..)..))))))))............--. ( -29.00) >DroSec_CAF1 52233 88 + 1 ------------AAACCCAUGAAAUCGGUGACACUGUCCGCCG----GAUGGCCAUGGCAGCAGUUUAAUUGCAGCGACUGUAGUGUCAUUUCCAUUUGAACC--U ------------.............(((((........)))))----(((((..((((((((((((..........))))))..))))))..)))))......--. ( -22.10) >DroSim_CAF1 55064 88 + 1 ------------AAACCCAUGAAAUCGGUGACACUGUCCGCCG----GAUGGCCAUGGCUGCAGUUUAAUUGCAGCGACUGUAGUGUCAUUUCCAUUUGAACC--U ------------..............((((((((((((.(((.----...)))....((((((((...)))))))))))...)))))))))............--. ( -26.20) >DroEre_CAF1 53063 100 + 1 CCAAAAAAAGAGGAACCCAUGAAAUCGGUGACACUGUCCGCCG----GAUGGCCAUGGCUGCGGUUUAAUUGCAGCGACUGUAGUGUCAUUUCGAUUUGAACC--U ...................(.(((((((((((((((((.(((.----...)))....((((((((...)))))))))))...))))))))..)))))).)...--. ( -30.40) >DroYak_CAF1 55278 101 + 1 CCAAA---AGAAAAACCCAUGAAAUCGGUGACACUGUCCGCCGAAUGGAUGGCCGUGGCUGCAGUUUAAUUGCAGCGACUGUAGUGUCAUUCCGAUUUGAACC--U .....---...........(.(((((((((((((((((((.....))))).((.((.((((((((...)))))))).)).)))))))))..))))))).)...--. ( -35.80) >DroPer_CAF1 124533 91 + 1 G---G---AGCGG----CAGGCGGCAGGCGACAGAGGCAGCGG----CAUCG-GAUGGCAGGAGUUUAAUUGCAGUGACUGUAAUGUCAUUUCCAUUUGAACCCCG (---(---.((.(----(..((..(.(....).)..)).)).)----).(((-((((((((........))))((((((......))))))..)))))))...)). ( -25.80) >consensus __________A_AAACCCAUGAAAUCGGUGACACUGUCCGCCG____GAUGGCCAUGGCUGCAGUUUAAUUGCAGCGACUGUAGUGUCAUUUCCAUUUGAACC__U ..........................((((((((((...((((......))))....((((((((...)))))))).....))))))))))............... (-19.91 = -20.42 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:25 2006