
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,204,656 – 3,204,762 |
| Length | 106 |
| Max. P | 0.708856 |

| Location | 3,204,656 – 3,204,762 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.48 |
| Mean single sequence MFE | -19.86 |
| Consensus MFE | -12.45 |
| Energy contribution | -12.34 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3204656 106 - 27905053 --AAAA-----AUGAGAAAA----AAAACCCAAAAACCGAGCGGAUAUUAUUGCGGCUCAAUAAAAUACUGGAUCGAGCCUUCGCAUGAAAUAAGGUUGUAAUAAAGGCCGCGU---AUC --....-----.........----................((((...((((((((((((................)))))(((....)))........)))))))...))))..---... ( -20.59) >DroGri_CAF1 24910 90 - 1 --AAAA-----AAGGG--A-----------------GAGAACGGAAAUUAUUGCAGCUCAAUAAAACAUGUAAGCAAACACUCAGGU-AAGGAAAAUUGUAAUAAGCGUCACAU---AGC --....-----.....--.-----------------.......((..(((((((((((((........))..)))....((....))-.........))))))))...))....---... ( -8.10) >DroSec_CAF1 24037 110 - 1 --AAAA-----ACGAAAAAAAAAAAAAACCCAAAAACCGAGCGGAUAUUAUUGCGGCUCAAUAAAAUACUGGAUCGAGCCAUCGCAUGGAAUAAGGUUGUAAUAAGGGCCGCGU---AUC --....-----.............................((((...((((((((((((................))))).((.....))........)))))))...))))..---... ( -21.99) >DroSim_CAF1 30891 105 - 1 --AAAA-----ACGAGAAAA-----AACCCAAAAAACCGAGCGGAUAUUAUUGCGGCUCAAUAAAGUACUGGAUCGAGCCAUCGCAUGGAAUAAGGUUGUAAUAAGGGCCGCGU---AUC --....-----.........-----...............((((...((((((((((((.((..(....)..)).))))).((.....))........)))))))...))))..---... ( -22.40) >DroEre_CAF1 23648 100 - 1 --AAAU-----GCGAAAA-------AAGGCCAAAAGCCGAGCGGAUAUUAUUGCGGCCCAAUAAAACACUGGACAGA---AUCGCAUGAAAUAAGGUUCUAAUAAGGGCUGCGU---AUC --..((-----((.....-------..(((.....)))..............(((((((.......(....)..(((---(((...........)))))).....)))))))))---)). ( -23.60) >DroMoj_CAF1 29442 102 - 1 AAAAAAUGUGUACGAGGAG-----------------CAAAACGGAAAUUAUUGCAGCUCAAUAAAACAUGUGGGCAAACAUGCAGGU-GAGUGAAAUUGUAAUAAGCGUCACGUACUAGC .........(((((.(..(-----------------(..........(((((((((((((......(((((......)))))....)-)))).....))))))))))..).))))).... ( -22.50) >consensus __AAAA_____ACGAGAAA______AA_CCCAAAAACCGAGCGGAUAUUAUUGCGGCUCAAUAAAACACUGGAUCGAGCCAUCGCAUGAAAUAAGGUUGUAAUAAGGGCCGCGU___AUC ........................................((((...(((((((((((.......((..(((......)))....)).......)))))))))))...))))........ (-12.45 = -12.34 + -0.11)

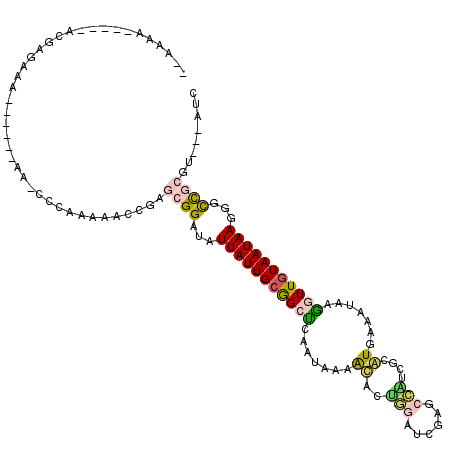
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:19 2006