
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,130,902 – 27,131,006 |
| Length | 104 |
| Max. P | 0.843605 |

| Location | 27,130,902 – 27,131,006 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -19.25 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
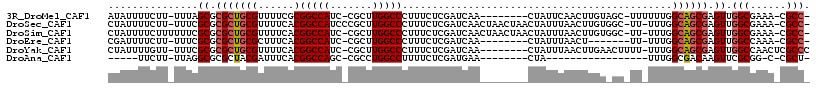
>3R_DroMel_CAF1 27130902 104 + 27905053 AUAUUUUCUU-UUUAGCGCGCUGCGUUUUCGCGGCCAUC-CGCUUGGCCCUUUCUCGAUCAA--------CUAUUCAACUUGUAGC-UUUUUUGGCAGCGAGUUGGCGAAA-CGCC- ..........-...........((((((.((((((((..-....))))).............--------.....((((((((.((-(.....))).))))))))))))))-))).- ( -34.50) >DroSec_CAF1 32763 112 + 1 CUAUUUUCUU-UUUCGCGCGCUGCGUUUUCACGGCCAUCCCGCUUGGCCCUUUCUCGAUCAACUAACUAACUAUUUAACUUGUGGC-UU-UUUGGCAGCGAGUUGGCGAAA-CGCC- ..........-...........(((((((...(((((.......))))).........................(((((((((.((-(.-...))).))))))))).))))-))).- ( -27.30) >DroSim_CAF1 35095 112 + 1 CUAUUUUCUUUUUUCGCGCGCUGCGUUUUCACGGCCAUC-CGCUUGGCCCUUUCUCGAUCAACUAACUAACUAUUUAACUUGUGGC-UU-UUUGGCAGCGAGUUGGCGAAA-CGCC- ......................(((((((...(((((..-....))))).........................(((((((((.((-(.-...))).))))))))).))))-))).- ( -27.80) >DroEre_CAF1 32825 97 + 1 CGAUUUUCUU-UUUCGCGCGCUGCGCUUUCACGGCCAUC-CGCUUGGCCCUUUCUCGAUCAA--------CUAUUUAACU-------UU-UUUGGCAGCGAGUUGGCCAAA-CGCC- ..........-....((.((((((........(((((..-....))))).............--------........(.-------..-...))))))).)).(((....-.)))- ( -22.30) >DroYak_CAF1 34370 106 + 1 CUAUUUUGUU-UUUCGCGCGCUGCGUUUUCACGGCCAUC-CGCUUGGCCCUUUCUCGAUCAA--------CUAUUUAACUUGAACUUUU-UUUGGCAGCGAGUUGGCCAACUCGCCC ..........-......(((((((........(((((..-....))))).........((((--------.........))))......-....)))))(((((....))))))).. ( -25.90) >DroAna_CAF1 35493 82 + 1 -----UUCUU-UUAGGCGCGCUACGAUUUCACGGCCAGC-CGCCUGGCCUUUUCUCGAUGAA--------CUA-----------------UUUGGCGACAAGUUCGCGG-C-CGCU- -----.....-...(((.(((..(((......((((((.-...)))))).....)))..(((--------((.-----------------..((....)))))))))))-)-)...- ( -28.20) >consensus CUAUUUUCUU_UUUCGCGCGCUGCGUUUUCACGGCCAUC_CGCUUGGCCCUUUCUCGAUCAA________CUAUUUAACUUGU_GC_UU_UUUGGCAGCGAGUUGGCGAAA_CGCC_ ...............((.(((((((......)(((((.......))))).............................................)))))).)).(((......))). (-19.25 = -19.78 + 0.53)


| Location | 27,130,902 – 27,131,006 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -21.45 |
| Energy contribution | -21.62 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
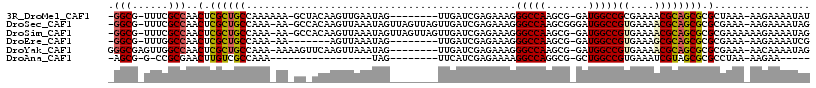
>3R_DroMel_CAF1 27130902 104 - 27905053 -GGCG-UUUCGCCAACUCGCUGCCAAAAAA-GCUACAAGUUGAAUAG--------UUGAUCGAGAAAGGGCCAAGCG-GAUGGCCGCGAAAACGCAGCGCGCUAAA-AAGAAAAUAU -.(((-((((((...((((((........)-))..(((.(.....).--------)))...)))....(((((....-..)))))))).))))))...........-.......... ( -27.60) >DroSec_CAF1 32763 112 - 1 -GGCG-UUUCGCCAACUCGCUGCCAAA-AA-GCCACAAGUUAAAUAGUUAGUUAGUUGAUCGAGAAAGGGCCAAGCGGGAUGGCCGUGAAAACGCAGCGCGCGAAA-AAGAAAAUAG -.(((-((..((((.(((((((((...-..-....(((.((((........)))))))..........)))..)))))).))))(((....))).)))))......-.......... ( -27.71) >DroSim_CAF1 35095 112 - 1 -GGCG-UUUCGCCAACUCGCUGCCAAA-AA-GCCACAAGUUAAAUAGUUAGUUAGUUGAUCGAGAAAGGGCCAAGCG-GAUGGCCGUGAAAACGCAGCGCGCGAAAAAAGAAAAUAG -....-((((((.....((((((....-..-.....................................(((((....-..)))))((....)))))))).))))))........... ( -26.60) >DroEre_CAF1 32825 97 - 1 -GGCG-UUUGGCCAACUCGCUGCCAAA-AA-------AGUUAAAUAG--------UUGAUCGAGAAAGGGCCAAGCG-GAUGGCCGUGAAAGCGCAGCGCGCGAAA-AAGAAAAUCG -.(((-((.(((((.(.(((((((...-..-------.(((((....--------)))))........)))..))))-).)))))((....))..)))))......-.......... ( -26.94) >DroYak_CAF1 34370 106 - 1 GGGCGAGUUGGCCAACUCGCUGCCAAA-AAAAGUUCAAGUUAAAUAG--------UUGAUCGAGAAAGGGCCAAGCG-GAUGGCCGUGAAAACGCAGCGCGCGAAA-AACAAAAUAG ..(((.((((((((.(.(((((((...-....(.((((.(.....).--------)))).).......)))..))))-).))))(((....))))))).)))....-.......... ( -30.94) >DroAna_CAF1 35493 82 - 1 -AGCG-G-CCGCGAACUUGUCGCCAAA-----------------UAG--------UUCAUCGAGAAAAGGCCAGGCG-GCUGGCCGUGAAAUCGUAGCGCGCCUAA-AAGAA----- -...(-(-(((((((((..........-----------------.))--------)))..(((.....((((((...-.))))))......)))..))).)))...-.....----- ( -28.50) >consensus _GGCG_UUUCGCCAACUCGCUGCCAAA_AA_GC_ACAAGUUAAAUAG________UUGAUCGAGAAAGGGCCAAGCG_GAUGGCCGUGAAAACGCAGCGCGCGAAA_AAGAAAAUAG .(((......)))..(.((((((.............................................(((((.......)))))((....)))))))).)................ (-21.45 = -21.62 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:14 2006