
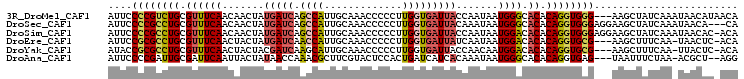
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,088,749 – 27,088,851 |
| Length | 102 |
| Max. P | 0.813332 |

| Location | 27,088,749 – 27,088,851 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -16.63 |
| Energy contribution | -16.91 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27088749 102 + 27905053 AUUCCCCGUCUGCGUUUCAACAACUAUGAUCAGCCAUUGCAAACCCCCUUGGUGAUUACCAAUAAUGGGCACACAGGUGGG---AAGCUAUCAAAUAACAUAACA ....(((..(((.((...........((((((.(((.............)))))))))(((....)))..)).)))..)))---..................... ( -20.72) >DroSec_CAF1 7661 102 + 1 AUUCCCCGCCUGCGUUUCAACAACUAUGAUCAGCCAUUGCAAACCCCCUUGGUGAUUACAAAUAAUGGGCACACAGGUGGGAGGAAGCUAUCAAAUAACA---CA .(((((((((((.((((((.......((((((.(((.............))))))))).......)))).)).)))))))).)))...............---.. ( -26.86) >DroSim_CAF1 7642 104 + 1 AUUCCCCGCCUGCGUUUCAACAACUAUGAUCAGCCAUUGCAAACCCCCUUGGUGAUUACCAAUAAUGGACACACAGGUGGGAGGAAGCUAUCAAAUAACAC-ACA .(((((((((((.((((((.......((((((.(((.............))))))))).......)))).)).)))))))).)))................-... ( -27.26) >DroEre_CAF1 7933 100 + 1 AUUCCGCGCCUGCGUUUCAACUACUAUGAUCAACCAUUGCAAACCCCCUUGGUGAUUAUCAAUAAUGGACACACAGGUGCG---AAGCUUUCAA-UAACUC-ACA ....((((((((.((((((......(((((((.(((.............))))))))))......)))).)).))))))))---..........-......-... ( -26.22) >DroYak_CAF1 7710 100 + 1 AUACCGCGCCUGCGUUUCAACUACUACGAUCAAGCAUUGCAAACCCCCUUGGUGAUUACCAACAAUGGACACACAGGUGCG---AAGCUUUCAA-UUACUC-ACA ....((((((((.((.((........(......)(((((.........(((((....)))))))))))).)).))))))))---..........-......-... ( -22.20) >DroAna_CAF1 7324 99 + 1 AUUCCCCGAUUGCGAUUCAAUUACUAUAACCAAACGCUUCGUACUCCACUGAUCAUCACAAAUAAUGGGCACACAGGUGAG---UAAUUUCUAA-ACGCU--AGG .....((....(((....(((((((...(((..(((...))).......((..(((........)))..))....))).))---))))).....-.))).--.)) ( -13.70) >consensus AUUCCCCGCCUGCGUUUCAACAACUAUGAUCAACCAUUGCAAACCCCCUUGGUGAUUACCAAUAAUGGACACACAGGUGGG___AAGCUAUCAA_UAACAC_ACA ....((((((((.((((((.......(((((.((((.............))))))))).......)))).)).))))))))........................ (-16.63 = -16.91 + 0.28)


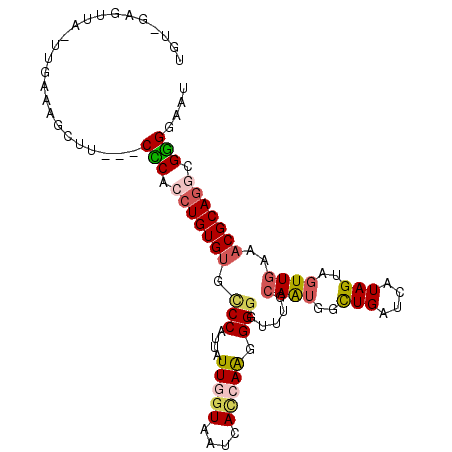
| Location | 27,088,749 – 27,088,851 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -19.81 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27088749 102 - 27905053 UGUUAUGUUAUUUGAUAGCUU---CCCACCUGUGUGCCCAUUAUUGGUAAUCACCAAGGGGGUUUGCAAUGGCUGAUCAUAGUUGUUGAAACGCAGACGGGGAAU .(((((........)))))((---(((..((((((((((....(((((....)))))..))))...(((..((((....))))..)))..))))))...))))). ( -34.10) >DroSec_CAF1 7661 102 - 1 UG---UGUUAUUUGAUAGCUUCCUCCCACCUGUGUGCCCAUUAUUUGUAAUCACCAAGGGGGUUUGCAAUGGCUGAUCAUAGUUGUUGAAACGCAGGCGGGGAAU ..---.(((((...)))))....((((.(((((((((((....((((.......)))).))))...(((..((((....))))..)))..))))))).))))... ( -31.30) >DroSim_CAF1 7642 104 - 1 UGU-GUGUUAUUUGAUAGCUUCCUCCCACCUGUGUGUCCAUUAUUGGUAAUCACCAAGGGGGUUUGCAAUGGCUGAUCAUAGUUGUUGAAACGCAGGCGGGGAAU ...-..(((((...)))))....((((.(((((((.(((....(((((....)))))..)))....(((..((((....))))..)))..))))))).))))... ( -32.30) >DroEre_CAF1 7933 100 - 1 UGU-GAGUUA-UUGAAAGCUU---CGCACCUGUGUGUCCAUUAUUGAUAAUCACCAAGGGGGUUUGCAAUGGUUGAUCAUAGUAGUUGAAACGCAGGCGCGGAAU ...-......-........((---(((.(((((((.((.(((((((...(((((((..(.......)..))).))))..))))))).)).))))))).))))).. ( -29.10) >DroYak_CAF1 7710 100 - 1 UGU-GAGUAA-UUGAAAGCUU---CGCACCUGUGUGUCCAUUGUUGGUAAUCACCAAGGGGGUUUGCAAUGCUUGAUCGUAGUAGUUGAAACGCAGGCGCGGUAU ...-((((..-......))))---(((.(((((((..((.((.(((((....))))).))))....(((((((.......)))).)))..))))))).))).... ( -26.20) >DroAna_CAF1 7324 99 - 1 CCU--AGCGU-UUAGAAAUUA---CUCACCUGUGUGCCCAUUAUUUGUGAUGAUCAGUGGAGUACGAAGCGUUUGGUUAUAGUAAUUGAAUCGCAAUCGGGGAAU ...--.....-..........---.((.(((((((((((((((((......))).))))).)))))..(((((..((((...))))..)).)))...)))))).. ( -22.30) >consensus UGU_GAGUUA_UUGAAAGCUU___CCCACCUGUGUGCCCAUUAUUGGUAAUCACCAAGGGGGUUUGCAAUGGCUGAUCAUAGUAGUUGAAACGCAGGCGGGGAAU ........................(((.(((((((.(((....(((((....))))).))).....((((..(((....)))..))))..))))))).))).... (-19.81 = -20.15 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:49:03 2006