
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,057,335 – 27,057,551 |
| Length | 216 |
| Max. P | 0.994326 |

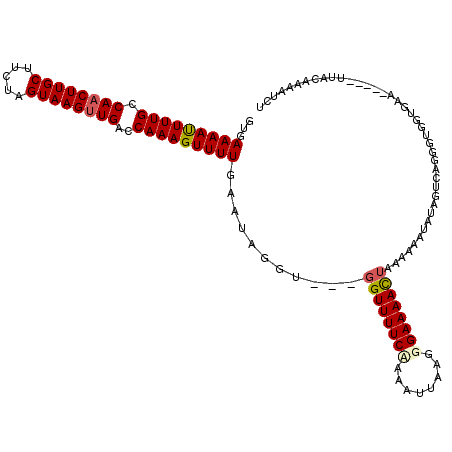
| Location | 27,057,335 – 27,057,452 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.44 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27057335 117 - 27905053 GUGAAAAUUUUGCCAACUUGCUUCUAGUAAGUUGACCAAAGUUUUGAAUAGGU---UGUUUUCUAAAUUAAGGGAAAACUAAAAAAUAUGGUCAGGGUGGUGAAUUAAAUUACAAAAUCU ((((....((..(((((((((.....))))))((((((..(((((...(((..---..((..((......))..))..))).))))).))))))...)))..)).....))))....... ( -26.70) >DroSec_CAF1 104575 112 - 1 GUGAAAAUUUUGCCAACUUGCUUCUAGUAAGUUGACCAAAGUUUUGAAUAGGU---GGUUUUCCAAAUUAAGGGAAAACUAAAAAAUAUAGUCAUGGUGGUGAA-----UUACAAAAUCU ((((((((((((.((((((((.....))))))))..))))))))........(---((((((((........)))))))))..........))))(((..((..-----...))..))). ( -26.40) >DroSim_CAF1 107372 112 - 1 GUGAAAAUUUUGCCAACUUGCUUCUAGUAAGUUGACCAAAGUUUUGAAUAGGU---GGUUUUCCAAAUUAAGGGAAAACUAAAAAAUAUAGUCAGGGUGGUGAA-----UUACAAAAACU ((((.........((((((((.....))))))))((((....((((((((..(---((((((((........))))))))).....)))..))))).))))...-----))))....... ( -26.80) >DroEre_CAF1 111863 103 - 1 GUAAAAAUUUUGCCAACUUGCUUCUAGUAAGUUGACCAAAGUUUUGAAUAGGUCGUGGUUUUCAAAAUUAGGGGAAAAUUAA-AAAUGUAAUCAGGGCGGU----------------UCU ...(((((((((.((((((((.....))))))))..)))))))))((((..(((.(((((..((...((((.......))))-...)).))))).))).))----------------)). ( -24.40) >DroYak_CAF1 107903 115 - 1 GUAAAAACUUUGCCAACUUGCUUCUAGUAAGCUGACCAAAGUUUUGAAAAGGUCGUGGUUUUCAAAAUUAAGGGAAAAUUAGGAAAUAUAGUCAGGGCGGUGAA-----UUAAACAAUGU ........((..((..(((((.....))))).........(((((((.....((.((((((((..........)))))))).)).......)))))))))..))-----........... ( -23.10) >consensus GUGAAAAUUUUGCCAACUUGCUUCUAGUAAGUUGACCAAAGUUUUGAAUAGGU___GGUUUUCAAAAUUAAGGGAAAACUAAAAAAUAUAGUCAGGGUGGUGAA_____UUACAAAAUCU ...(((((((((.((((((((.....))))))))..)))))))))...........((((((((........))))))))........................................ (-19.26 = -19.58 + 0.32)


| Location | 27,057,375 – 27,057,492 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.85 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -22.64 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27057375 117 - 27905053 UAAUAUUAAUAUCAGCCUAUAUGUCAUUUGUACUAUCUCUGUGAAAAUUUUGCCAACUUGCUUCUAGUAAGUUGACCAAAGUUUUGAAUAGGU---UGUUUUCUAAAUUAAGGGAAAACU .............(((((((..........(((.......)))(((((((((.((((((((.....))))))))..)))))))))..))))))---)(((((((........))))))). ( -27.50) >DroSec_CAF1 104610 117 - 1 UAAUAUUAACAUCAGCCUAUAUGCCAUUUGUACUAUCUCUGUGAAAAUUUUGCCAACUUGCUUCUAGUAAGUUGACCAAAGUUUUGAAUAGGU---GGUUUUCCAAAUUAAGGGAAAACU ..............((((((..........(((.......)))(((((((((.((((((((.....))))))))..)))))))))..))))))---((((((((........)))))))) ( -29.50) >DroSim_CAF1 107407 117 - 1 UAAUAUUAACAUCAGCCUAUAUGCCAUUUCUUCUAUCUCUGUGAAAAUUUUGCCAACUUGCUUCUAGUAAGUUGACCAAAGUUUUGAAUAGGU---GGUUUUCCAAAUUAAGGGAAAACU ..............((((((....(((.............)))(((((((((.((((((((.....))))))))..)))))))))..))))))---((((((((........)))))))) ( -28.62) >DroEre_CAF1 111886 120 - 1 UAAUAUAAAAACCAGCCUAGAAACCAUUUGUACUAUCUCUGUAAAAAUUUUGCCAACUUGCUUCUAGUAAGUUGACCAAAGUUUUGAAUAGGUCGUGGUUUUCAAAAUUAGGGGAAAAUU ...........((..((((((((((((.....((((.((....(((((((((.((((((((.....))))))))..)))))))))))))))...))))))))......))))))...... ( -26.70) >DroYak_CAF1 107938 120 - 1 UAAUAUUAACACCAGCCUAGAAACCAUUCGUACUAUCUCUGUAAAAACUUUGCCAACUUGCUUCUAGUAAGCUGACCAAAGUUUUGAAAAGGUCGUGGUUUUCAAAAUUAAGGGAAAAUU .........(((..((((............(((.......)))(((((((((.((.(((((.....))))).))..)))))))))....)))).)))((((((..........)))))). ( -21.20) >consensus UAAUAUUAACAUCAGCCUAUAUGCCAUUUGUACUAUCUCUGUGAAAAUUUUGCCAACUUGCUUCUAGUAAGUUGACCAAAGUUUUGAAUAGGU___GGUUUUCAAAAUUAAGGGAAAACU ..............((((((..........(((.......)))(((((((((.((((((((.....))))))))..)))))))))..))))))....(((((((........))))))). (-22.64 = -23.32 + 0.68)



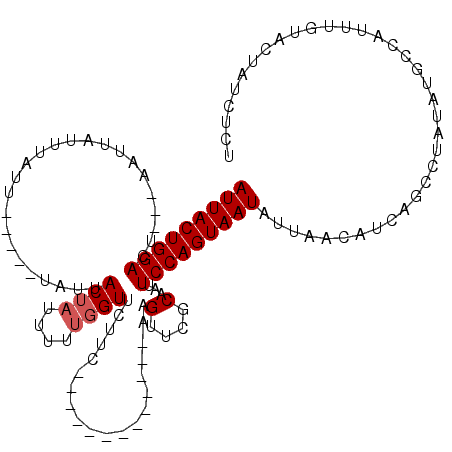
| Location | 27,057,452 – 27,057,551 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.04 |
| Mean single sequence MFE | -16.35 |
| Consensus MFE | -8.84 |
| Energy contribution | -9.24 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27057452 99 - 27905053 AUUACUGGAGU---AAUUAUUUAUU-----UAUUACUAUUUUAGGUUCUUC-------------AAGUUCGCAAUUCCAGUAAUAUUAAUAUCAGCCUAUAUGUCAUUUGUACUAUCUCU (((((((((((---...........-----....(((......))).....-------------.........))))))))))).................................... ( -12.15) >DroSec_CAF1 104687 99 - 1 AUUACUGGACU---AAUUAUUCAUU-----UAUUACUAUUUUUGGUUCUCC-------------AAGUUCGCAUUUCCAGUAAUAUUAACAUCAGCCUAUAUGCCAUUUGUACUAUCUCU (((((((((..---...........-----...........((((....))-------------))(....)...)))))))))....(((...((......))....)))......... ( -13.30) >DroSim_CAF1 107484 99 - 1 AUUACUGGACU---AAUUAUUCAUU-----UAUUACUAUUUUUGGUUCUCC-------------AAGUUCGCAUUUCCAGUAAUAUUAACAUCAGCCUAUAUGCCAUUUCUUCUAUCUCU (((((((((..---...........-----...........((((....))-------------))(....)...))))))))).................................... ( -12.40) >DroEre_CAF1 111966 119 - 1 AUUACUGGAGUGCAACUUAUUUAUUACUUGUAAAACUAUUUUUGGUUCUUCCUUCA-CCACUCAAAGUUCGCUAUUCCAGUAAUAUAAAAACCAGCCUAGAAACCAUUUGUACUAUCUCU ((((((((((((((((((.(((((.....)))))........((((.........)-)))....))))).)).))))))))))).................................... ( -21.40) >DroYak_CAF1 108018 120 - 1 AUUACUGGAUAGGAACUUCUUUAGUACUUAUAAAACUAUUUUUCGUUCUUCCUUAAGCUACUCAAAGUUCGCUAUUCCAGUAAUAUUAACACCAGCCUAGAAACCAUUCGUACUAUCUCU ((((((((((((((((((....((((((((...(((........)))......)))).))))..)))))).))).))))))))).................................... ( -22.50) >consensus AUUACUGGACU___AAUUAUUUAUU_____UAUUACUAUUUUUGGUUCUUC_____________AAGUUCGCAAUUCCAGUAAUAUUAACAUCAGCCUAUAUGCCAUUUGUACUAUCUCU (((((((((.........................((((....))))....................(....)...))))))))).................................... ( -8.84 = -9.24 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:48:54 2006