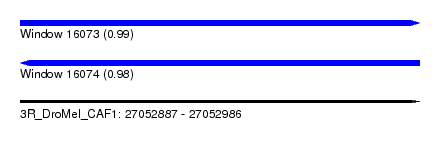
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,052,887 – 27,052,986 |
| Length | 99 |
| Max. P | 0.989325 |

| Location | 27,052,887 – 27,052,986 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.46 |
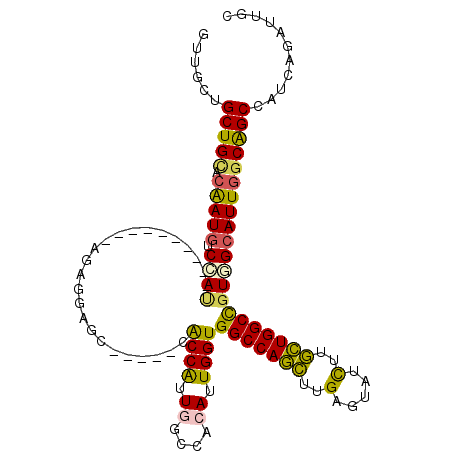
| Mean single sequence MFE | -36.75 |
| Consensus MFE | -21.36 |
| Energy contribution | -21.03 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27052887 99 + 27905053 GCAAUCUGAUGGCUGCUAAUGCCACAGCCAGCAAGAUACUCAAGCUGGCCACCAAUGUGGCCAAUGGUG-----GCUCCUCU----------AUGGACAUUGUGCAGCAGCAAC ((.((((..((((((.........))))))...))))......(((((((((((.((....)).)))))-----))(((...----------..))).......)))).))... ( -36.70) >DroEre_CAF1 107429 99 + 1 GUAAUCUGAUGGCUGCCAAUGCCACAGCCAGCAAGAUACUCAAGCUGGCCACCAAUGUGGCCAAUGGUG-----GCUCCACU----------AUGGACAUUGUGCAGCAGCAAC ......((...(((((((((((((((((((((..(.....)..))))))......)))))(((.(((((-----....))))----------)))).))))).)))))..)).. ( -39.00) >DroWil_CAF1 108613 114 + 1 GCAAUUUAAUGGCUGCCAAUGCAACGGCCAAUAAGAUCCUCAAAUUGGCCACCAAUGUGGCAAAUGGCGUUGUCGCUGCUCCACCGCAUGUGGUGGAUAUUGUUCAGCAGCACA ...........(((((....(((..(((((((..(.....)..)))))))......(((((((......))))))))))(((((((....))))))).........)))))... ( -40.00) >DroYak_CAF1 103424 99 + 1 GCAAUCUGAUGGCUGCCAAUGCUACAGCCAGCAAGAUACUCAAGCUGGCCACCAAUGUGGCCAACGGUG-----GCUCCUCU----------AUGGACAUUGUGCAGCAGCAAC ......((...((((((((((.....((((.(..(.....)..(.(((((((....))))))).)).))-----))(((...----------..)))))))).)))))..)).. ( -37.00) >DroMoj_CAF1 133198 88 + 1 --AAUUUAAUAGCCGCCAAUGCCACGGCCAACAAAAUACUCAAGUUGGCCACCAAUGUGGCAAACGGCG------------------------UGGACAUUGCACAGCAACACA --.........((((....(((((((((((((...........)))))))......))))))..))))(------------------------((....((((...))))))). ( -29.00) >DroAna_CAF1 110498 99 + 1 GCAAUCUUAUGGCUGCCAAUGCAACGGCCAGCAAGAUUCUCAAGCUGGCCACCAAUGUGGCCAACGGCG-----GCUCCGCC----------GUCGACAUCGUACAGCACCAGC ((((((((.((((((.........))))))..)))))).....(((((((((....))))))..(((((-----((...)))----------)))).........)))....)) ( -38.80) >consensus GCAAUCUGAUGGCUGCCAAUGCCACAGCCAGCAAGAUACUCAAGCUGGCCACCAAUGUGGCCAACGGCG_____GCUCCUCU__________AUGGACAUUGUGCAGCAGCAAC ...........(((((((((((((..((((((...........))))))(((....)))((.....)).........................))).)))))....)))))... (-21.36 = -21.03 + -0.33)

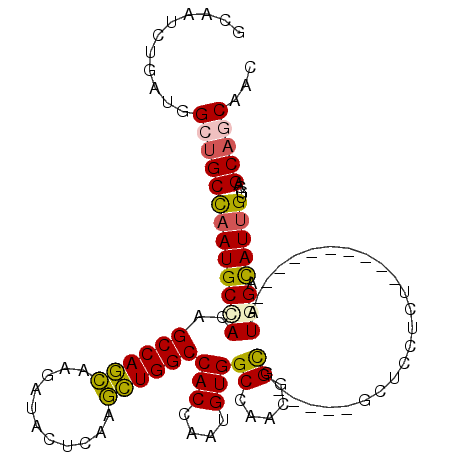
| Location | 27,052,887 – 27,052,986 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.46 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -31.49 |
| Energy contribution | -30.43 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.39 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27052887 99 - 27905053 GUUGCUGCUGCACAAUGUCCAU----------AGAGGAGC-----CACCAUUGGCCACAUUGGUGGCCAGCUUGAGUAUCUUGCUGGCUGUGGCAUUAGCAGCCAUCAGAUUGC ...(((((((....(((.((((----------((....((-----(((((.((....)).)))))))((((..(.....)..)))).))))))))))))))))........... ( -40.90) >DroEre_CAF1 107429 99 - 1 GUUGCUGCUGCACAAUGUCCAU----------AGUGGAGC-----CACCAUUGGCCACAUUGGUGGCCAGCUUGAGUAUCUUGCUGGCUGUGGCAUUGGCAGCCAUCAGAUUAC .(((..(((((.(((((.((((----------(((...((-----(((((.((....)).)))))))((((..(.....)..)))))))))))))))))))))...)))..... ( -46.80) >DroWil_CAF1 108613 114 - 1 UGUGCUGCUGAACAAUAUCCACCACAUGCGGUGGAGCAGCGACAACGCCAUUUGCCACAUUGGUGGCCAAUUUGAGGAUCUUAUUGGCCGUUGCAUUGGCAGCCAUUAAAUUGC ((.(((((..(......((((((......))))))((((((....(((((..........)))))((((((..(.....)..)))))))))))).)..)))))))......... ( -45.40) >DroYak_CAF1 103424 99 - 1 GUUGCUGCUGCACAAUGUCCAU----------AGAGGAGC-----CACCGUUGGCCACAUUGGUGGCCAGCUUGAGUAUCUUGCUGGCUGUAGCAUUGGCAGCCAUCAGAUUGC .(((..(((((.((((((((..----------...))(((-----(...(((((((((....)))))))))...(((.....)))))))...)))))))))))...)))..... ( -39.20) >DroMoj_CAF1 133198 88 - 1 UGUGUUGCUGUGCAAUGUCCA------------------------CGCCGUUUGCCACAUUGGUGGCCAACUUGAGUAUUUUGUUGGCCGUGGCAUUGGCGGCUAUUAAAUU-- ......(((((.(((((.(((------------------------(.......(((.....)))(((((((...........))))))))))))))))))))).........-- ( -33.80) >DroAna_CAF1 110498 99 - 1 GCUGGUGCUGUACGAUGUCGAC----------GGCGGAGC-----CGCCGUUGGCCACAUUGGUGGCCAGCUUGAGAAUCUUGCUGGCCGUUGCAUUGGCAGCCAUAAGAUUGC ((..((((....((((((((((----------((((....-----))))))))...))))))..(((((((..(.....)..)))))))...))))..)).............. ( -48.20) >consensus GUUGCUGCUGCACAAUGUCCAU__________AGAGGAGC_____CACCAUUGGCCACAUUGGUGGCCAGCUUGAGUAUCUUGCUGGCCGUGGCAUUGGCAGCCAUCAGAUUGC ......(((((.(((((.((((........................((((.((....)).))))(((((((..(.....)..)))))))))))))))))))))........... (-31.49 = -30.43 + -1.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:48:47 2006