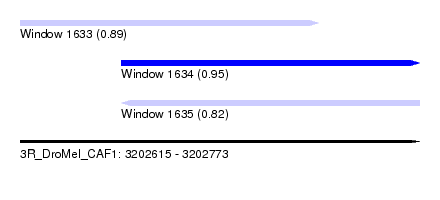
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,202,615 – 3,202,773 |
| Length | 158 |
| Max. P | 0.950441 |

| Location | 3,202,615 – 3,202,733 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -26.24 |
| Energy contribution | -26.80 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3202615 118 + 27905053 GUUGUAAACGUAAGUGGUAUGUUACUCAUGCAGACAAUCAACACUUGUCAUGAAAUGGCACCAUUUCCGCAACCGAAGUACCCUGUGUGUUCAUACACAUGUCGGGUGGUUUGUUUUU (((((....((((((((((((.....))))).(.....)..)))))).)..((((((....)))))).)))))((((.(((((.((((((....))))))...))))).))))..... ( -35.30) >DroSec_CAF1 22049 113 + 1 AUUGUAAACGUAAGUGGCAUGA-----GUAAAGACCAUCAACACUUGUCAUGAAAUGGCACCAUUUCCGCAACCUAAGUACCCUGUGUGUUCAUACACAUGUCGGGUGCUUUGUUUUU .((((....(((((((..(((.-----((....)))))...)))))).)..((((((....)))))).))))...((((((((.((((((....))))))...))))))))....... ( -31.10) >DroSim_CAF1 28890 113 + 1 AUUGCUAACGUAAGUGGCAUGA-----GUAAAGACAAUCAACACUUGUCAUGAAAUGGCACCAUUUCCGCAACCUAAGUACCCUGUGUGUUCAUACACAUGUCGGGUGCUUUGUUUUU ..(((((.(....))))))...-----((...(((((.......)))))..((((((....)))))).))(((..((((((((.((((((....))))))...)))))))).)))... ( -33.60) >DroEre_CAF1 21656 113 + 1 AUUGUUAACGUAUGUGGCAUGA-----GUAAAGGCAAUCAACACUUGUCAUGAAAUGGCACCAUUUCCGCAACCUAAGUACCCUGUGUGUUCAUACACAUUUCGGGCGCUUUGUUUUU ............((((((((((-----...(((..........))).)))))(((((....))))))))))((..(((..(((.((((((....))))))...)))..))).)).... ( -27.00) >consensus AUUGUAAACGUAAGUGGCAUGA_____GUAAAGACAAUCAACACUUGUCAUGAAAUGGCACCAUUUCCGCAACCUAAGUACCCUGUGUGUUCAUACACAUGUCGGGUGCUUUGUUUUU .............(((................(((((.......)))))..((((((....)))))))))(((..((((((((.((((((....))))))...)))))))).)))... (-26.24 = -26.80 + 0.56)

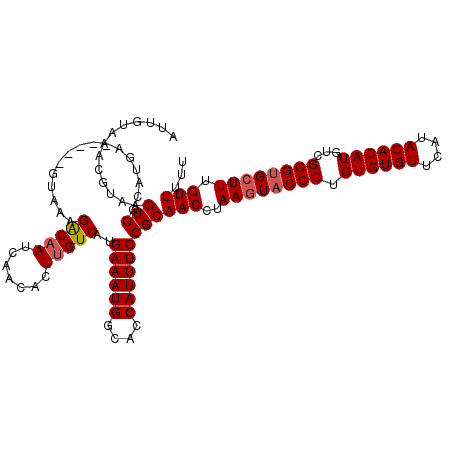
| Location | 3,202,655 – 3,202,773 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 97.46 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -29.38 |
| Energy contribution | -29.88 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3202655 118 + 27905053 ACACUUGUCAUGAAAUGGCACCAUUUCCGCAACCGAAGUACCCUGUGUGUUCAUACACAUGUCGGGUGGUUUGUUUUUGCUGAACAUCUUUAUCCGAAACGAUAAUUUUAUUGGACAU .....((((..((((((....)))))).((((.((((.(((((.((((((....))))))...))))).))))...))))...................(((((....))))))))). ( -30.90) >DroSec_CAF1 22084 118 + 1 ACACUUGUCAUGAAAUGGCACCAUUUCCGCAACCUAAGUACCCUGUGUGUUCAUACACAUGUCGGGUGCUUUGUUUUUGCUGAACAUCUUUAUCCGAAACGAUAAUUUUAUUGGACAU .....((((..((((((....)))))).((((...((((((((.((((((....))))))...)))))))).....))))...................(((((....))))))))). ( -31.80) >DroSim_CAF1 28925 118 + 1 ACACUUGUCAUGAAAUGGCACCAUUUCCGCAACCUAAGUACCCUGUGUGUUCAUACACAUGUCGGGUGCUUUGUUUUUGCUGAACAUCUUUAUCCGAAACGAUAAUUUUAUUGGACAU .....((((..((((((....)))))).((((...((((((((.((((((....))))))...)))))))).....))))...................(((((....))))))))). ( -31.80) >DroEre_CAF1 21691 118 + 1 ACACUUGUCAUGAAAUGGCACCAUUUCCGCAACCUAAGUACCCUGUGUGUUCAUACACAUUUCGGGCGCUUUGUUUUUGCUGAACAGCUUUAUCUGAAACGAUAAUUUUAUUGGACAU .....((((..((((((....)))))).((.....(((..(((.((((((....))))))...)))..)))(((((.....))))))).(((((......)))))........)))). ( -29.40) >consensus ACACUUGUCAUGAAAUGGCACCAUUUCCGCAACCUAAGUACCCUGUGUGUUCAUACACAUGUCGGGUGCUUUGUUUUUGCUGAACAUCUUUAUCCGAAACGAUAAUUUUAUUGGACAU .....((((..((((((....)))))).((((...((((((((.((((((....))))))...)))))))).....)))).........(((((......)))))........)))). (-29.38 = -29.88 + 0.50)

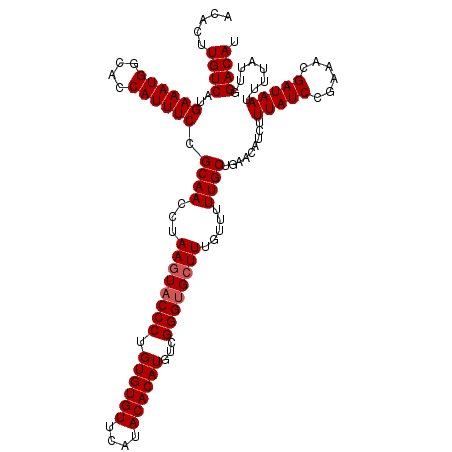
| Location | 3,202,655 – 3,202,773 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 97.46 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -29.11 |
| Energy contribution | -29.42 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3202655 118 - 27905053 AUGUCCAAUAAAAUUAUCGUUUCGGAUAAAGAUGUUCAGCAAAAACAAACCACCCGACAUGUGUAUGAACACACAGGGUACUUCGGUUGCGGAAAUGGUGCCAUUUCAUGACAAGUGU .((((.........((((((((((((((....))))).((((.........((((....(((((....)))))..)))).(....))))).))))))))).........))))..... ( -28.07) >DroSec_CAF1 22084 118 - 1 AUGUCCAAUAAAAUUAUCGUUUCGGAUAAAGAUGUUCAGCAAAAACAAAGCACCCGACAUGUGUAUGAACACACAGGGUACUUAGGUUGCGGAAAUGGUGCCAUUUCAUGACAAGUGU .((((.........((((((((((((((....))))).((((...(.(((.((((....(((((....)))))..)))).))).).)))).))))))))).........))))..... ( -30.97) >DroSim_CAF1 28925 118 - 1 AUGUCCAAUAAAAUUAUCGUUUCGGAUAAAGAUGUUCAGCAAAAACAAAGCACCCGACAUGUGUAUGAACACACAGGGUACUUAGGUUGCGGAAAUGGUGCCAUUUCAUGACAAGUGU .((((.........((((((((((((((....))))).((((...(.(((.((((....(((((....)))))..)))).))).).)))).))))))))).........))))..... ( -30.97) >DroEre_CAF1 21691 118 - 1 AUGUCCAAUAAAAUUAUCGUUUCAGAUAAAGCUGUUCAGCAAAAACAAAGCGCCCGAAAUGUGUAUGAACACACAGGGUACUUAGGUUGCGGAAAUGGUGCCAUUUCAUGACAAGUGU .............(((((......))))).(((..(((((((...(.(((..(((....(((((....)))))..)))..))).).)))).((((((....)))))).)))..))).. ( -31.10) >consensus AUGUCCAAUAAAAUUAUCGUUUCGGAUAAAGAUGUUCAGCAAAAACAAAGCACCCGACAUGUGUAUGAACACACAGGGUACUUAGGUUGCGGAAAUGGUGCCAUUUCAUGACAAGUGU .((((.........((((((((((((((....))))).((((...(.(((.((((....(((((....)))))..)))).))).).)))).))))))))).........))))..... (-29.11 = -29.42 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:16 2006