| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,010,466 – 27,010,571 |
| Length | 105 |
| Max. P | 0.897821 |

| Location | 27,010,466 – 27,010,571 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -20.34 |
| Energy contribution | -19.04 |
| Covariance contribution | -1.30 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
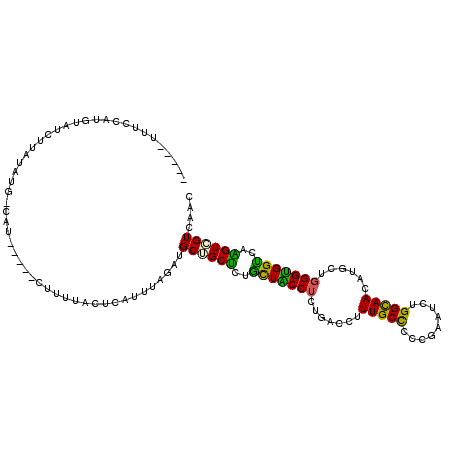
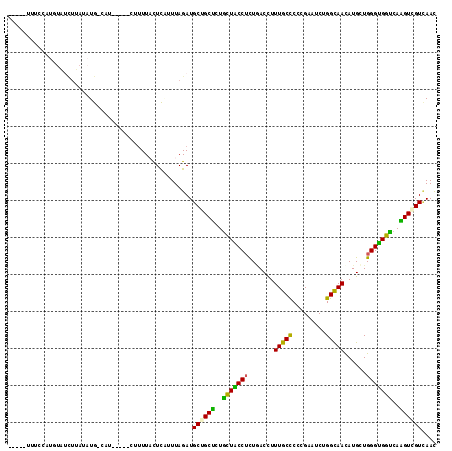
>3R_DroMel_CAF1 27010466 105 - 27905053 -----UUUCCAUGUAUCUUAUAUG-CAU-----CUUUUACUCAUUUAGAUGCUGCUCUGCUACCUCUUACCUUUGCCCCCGAGUCUGGUAAUAUGCUGGGUGGUCAAGUCGUCAAU -----..................(-(((-----((...........)))))).(((..(((((((.......(((((.........)))))......)))))))..)))....... ( -20.22) >DroGri_CAF1 79869 100 - 1 -----UUUGAU---------UUUG-CCUUCCUUCCUUUCCUC-UUUAGAUGCUGCCCUACUGCCGCUGACCUUUGCCCCGGAGUCGGGCAAUAUGUUGGGCGGUCAGGUGGUGAAC -----......---------((..-((......(((..((.(-(..(.(((.(((((.(((.(((..(.......)..)))))).))))).))).)..)).))..))).))..)). ( -28.40) >DroSec_CAF1 57860 103 - 1 -----UCUCCCUGUAUCUUAUAUG-CAU-------UUUACUCAUUUAGAUGCUGCUCUGCUACCUCUAACCUUUGCCCCCGAGUCUGGUAACAUGCUGGGUGGCCAAGUCGUCAAU -----.......((((((...(((-...-------......)))..))))))..................(((.((((((.(((.((....)).)))))).))).)))........ ( -21.00) >DroEre_CAF1 60704 105 - 1 -----UUUCGAUGUGUCUUAUAUG-UAU-----CUUUUACUCCAUUAGAUGCGGCUUUGCUACCUCUGACUUUUGCCCCCGAAUCUGGCAACAUGCUGGGUGGUCAAGUCGUCAAU -----....(((((((((.....(-((.-----....)))......))))))(((((.(((((((..(....(((((.........)))))....).))))))).))))))))... ( -28.70) >DroYak_CAF1 60254 110 - 1 UGUCAUUUCCAUGUGCCUUAUAUG-CAU-----UUUUUACUCAUUUAGAUGCGGCUCUGCUACCUCUGACCUUUGCCCCCGAAUCGGGCAACAUGCUGGGUGGCCAAGUCGUCAAC ((.(...................(-(((-----((...........))))))((((..(((((((..(....((((((.......))))))....).)))))))..))))).)).. ( -29.50) >DroAna_CAF1 62383 96 - 1 --------------UUUUUAUUUGACAU-----U-GUUUUUUGAUUAGAUGCUGCUCUGUUGCCUUUGACCUUUGCUCCCGAAUCUGGCAACAUGUUGGGUGGUCAGGUGGUUAAC --------------....(((((((((.-----.-......)).))))))).......(((((((((((((......(((((...((....))..))))).))))))).)).)))) ( -22.80) >consensus _____UUUCCAUGUAUCUUAUAUG_CAU_____CUUUUACUCAUUUAGAUGCUGCUCUGCUACCUCUGACCUUUGCCCCCGAAUCUGGCAACAUGCUGGGUGGUCAAGUCGUCAAC ..................................................((((((..(((((((.......(((((.........)))))......)))))))..)))))).... (-20.34 = -19.04 + -1.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:48:30 2006