
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,998,450 – 26,998,557 |
| Length | 107 |
| Max. P | 0.749213 |

| Location | 26,998,450 – 26,998,557 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -14.56 |
| Energy contribution | -15.17 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26998450 107 + 27905053 CAUUUUAAGUGCACACA------UAUAUCGCCAUUGUAGAUAUCUGUCUAUCCUUCGGAUA------UAUAUCUGCUUAUGGCCUCGAUAUGGCGCACAUUAUCGUUAAAUGCCAGCGG ........((((.(.((------(((...(((((.((((((((.(((((.......)))))------.))))))))..)))))....)))))).)))).....((((.......)))). ( -33.10) >DroVir_CAF1 70285 83 + 1 CAUUGUGGGCGG-----------------ACCAUUUUAUAU--U-GG------CUC---UC------AAUGUCUGUUUAUGGCCUCGAUAUG-CGCACAUUAUCGUUAAAUGCCAACAA ..((((.(((((-----------------.((((...((((--(-(.------...---.)------)))))......)))))).(((((((-....)).)))))......))).)))) ( -18.90) >DroSec_CAF1 45816 107 + 1 CAUUUUAAGUGCACACA------UAUAUCGCCAUUGUAGAUAUCUGUCUACCCUUCGGAUA------UAUAUCUGCUUAUGGCCUCGAUAUGGCGCACAUUAUCGUUAAAUGCCAGCGG ........((((.(.((------(((...(((((.((((((((.(((((.......)))))------.))))))))..)))))....)))))).)))).....((((.......)))). ( -33.10) >DroSim_CAF1 45889 107 + 1 CAUUUUAAGUGCACACA------UAUAUCGCCAUUGUAGAUAUCUGUCUACCCUUCGGAUA------UAUAUCUGCUUAUGGCCUCGAUAUGGCGCACAUUAUCGUUAAAUGCCAGCGG ........((((.(.((------(((...(((((.((((((((.(((((.......)))))------.))))))))..)))))....)))))).)))).....((((.......)))). ( -33.10) >DroEre_CAF1 48085 113 + 1 CAUUUUAAGUGCACACAUACUUGUAUAUGGGCAUUGUAGAUAUCCGUCCAUCCCUCGGAUA------UAUAUCUGCUUAUGGCCUCGAUAUGGCGCACAUUAUCGUUAAAUGCCAGCGG ........((((.(.((((.(((.....((.(((.((((((((..((((.......)))).------.))))))))..))).)).)))))))).)))).....((((.......)))). ( -31.00) >DroYak_CAF1 46489 115 + 1 CAUUUUAAGUGCACACACA----UAUAUUGGCAUUGUAGAUAUCCGACCAUCCUUCGGAUAUAAGUAUAUAUCUGCUUAUGGCCUCGAUAUGGCGCACAUUAUCGUUAAAUGCCAGCGG ........(((....))).----....((((((((...(((((((((.......)))))((((((((......))))))))(((.......)))......))))....))))))))... ( -29.50) >consensus CAUUUUAAGUGCACACA______UAUAUCGCCAUUGUAGAUAUCUGUCUAUCCUUCGGAUA______UAUAUCUGCUUAUGGCCUCGAUAUGGCGCACAUUAUCGUUAAAUGCCAGCGG ..........((.......................(((((((((((.........))))..........)))))))...((((..(((((((.....)).)))))......)))))).. (-14.56 = -15.17 + 0.61)

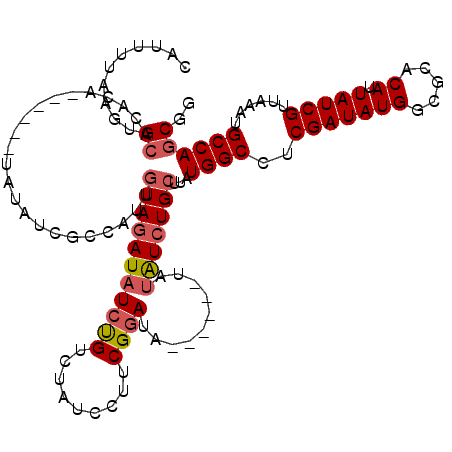

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:48:26 2006