
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,993,212 – 26,993,358 |
| Length | 146 |
| Max. P | 0.996212 |

| Location | 26,993,212 – 26,993,318 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -20.48 |
| Consensus MFE | -18.92 |
| Energy contribution | -18.84 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26993212 106 + 27905053 UUAAAUUAUAAAUAAUUGCCAUAUGAAGAGAUGGCAUUUAAAAUGGAAGAAAAUUAAGUUUAGCAGACUCUGCCAAAGGAAAUUAAGCCCGAAACAACUUUCCAUA ............(((.((((((........)))))).)))..(((((((.........(((.((((...)))).)))((.........))........))))))). ( -18.70) >DroSec_CAF1 40715 106 + 1 UUAAAUUAUAAAUAAUUGCCAUAUGAAGAGCUGGCAUUUAAAAUGCAAGAAAAUUAAGUUUAGCAGACUCUGCCAAAGGAAAUUAAGCCAGAAACAACUUGCCAUA ............(((.(((((..........))))).)))....(((((.........(((.((((...)))).)))((........))........))))).... ( -16.60) >DroSim_CAF1 40779 106 + 1 UUAAAUUAUAAAUAAUUGCCAUAUGAAGAGCAGGCAUUUAAAAUGCAAGAAAAUUAAGUUUGGCAGACUCUGCCAAAGGAAAUUAAGCCAGAAACAACUUGCCAUA ............(((.((((............)))).)))....(((((.........((((((((...))))))))((........))........))))).... ( -22.00) >DroEre_CAF1 43000 106 + 1 UUAAAUUAUAAAUAAUUGCCAUCUGAAGAGCUGGCAUUUAAAAUGCAAGAAAAUUAAGUUUGGCAGACUCUGCCAAAGGAAAUUGAGCCAGCAACUGCUUGCCAUA ............(((.(((((.((....)).))))).)))....(((((.........((((((((...))))))))((........))........))))).... ( -25.10) >DroYak_CAF1 41457 106 + 1 UUAAAUUAUAAAUAAUUGCCAUAUGCAAAGCCGGCAUUUAAAAUGCAAGAAAAUUAAGUUUGGCAGACUUUGCCAAAGGAUAUUUGGCCAAAAACUACUUGCCAUA ............(((.((((............)))).)))....(((((.........((((((((...))))))))((........))........))))).... ( -20.00) >consensus UUAAAUUAUAAAUAAUUGCCAUAUGAAGAGCUGGCAUUUAAAAUGCAAGAAAAUUAAGUUUGGCAGACUCUGCCAAAGGAAAUUAAGCCAGAAACAACUUGCCAUA ............(((.((((............)))).)))....(((((.........((((((((...))))))))((........))........))))).... (-18.92 = -18.84 + -0.08)



| Location | 26,993,212 – 26,993,318 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26993212 106 - 27905053 UAUGGAAAGUUGUUUCGGGCUUAAUUUCCUUUGGCAGAGUCUGCUAAACUUAAUUUUCUUCCAUUUUAAAUGCCAUCUCUUCAUAUGGCAAUUAUUUAUAAUUUAA ...(((((((((....(((.......)))((((((((...))))))))..))))))))).......(((.((((((........)))))).)))............ ( -20.40) >DroSec_CAF1 40715 106 - 1 UAUGGCAAGUUGUUUCUGGCUUAAUUUCCUUUGGCAGAGUCUGCUAAACUUAAUUUUCUUGCAUUUUAAAUGCCAGCUCUUCAUAUGGCAAUUAUUUAUAAUUUAA ....(((((........((........))((((((((...)))))))).........)))))....(((.(((((..........))))).)))............ ( -18.90) >DroSim_CAF1 40779 106 - 1 UAUGGCAAGUUGUUUCUGGCUUAAUUUCCUUUGGCAGAGUCUGCCAAACUUAAUUUUCUUGCAUUUUAAAUGCCUGCUCUUCAUAUGGCAAUUAUUUAUAAUUUAA ....(((((........((........))((((((((...)))))))).........)))))(((.((((((..((((........))))..)))))).))).... ( -20.20) >DroEre_CAF1 43000 106 - 1 UAUGGCAAGCAGUUGCUGGCUCAAUUUCCUUUGGCAGAGUCUGCCAAACUUAAUUUUCUUGCAUUUUAAAUGCCAGCUCUUCAGAUGGCAAUUAUUUAUAAUUUAA ..((((.(((....))).)).))......((((((((...))))))))..................(((.(((((.((....)).))))).)))............ ( -22.60) >DroYak_CAF1 41457 106 - 1 UAUGGCAAGUAGUUUUUGGCCAAAUAUCCUUUGGCAAAGUCUGCCAAACUUAAUUUUCUUGCAUUUUAAAUGCCGGCUUUGCAUAUGGCAAUUAUUUAUAAUUUAA ..((.((.((.((....((((........(((((((.....)))))))............((((.....)))).))))..)))).)).))................ ( -20.40) >consensus UAUGGCAAGUUGUUUCUGGCUUAAUUUCCUUUGGCAGAGUCUGCCAAACUUAAUUUUCUUGCAUUUUAAAUGCCAGCUCUUCAUAUGGCAAUUAUUUAUAAUUUAA ....(((((........((........))((((((((...)))))))).........)))))....(((.(((((..........))))).)))............ (-17.40 = -17.64 + 0.24)

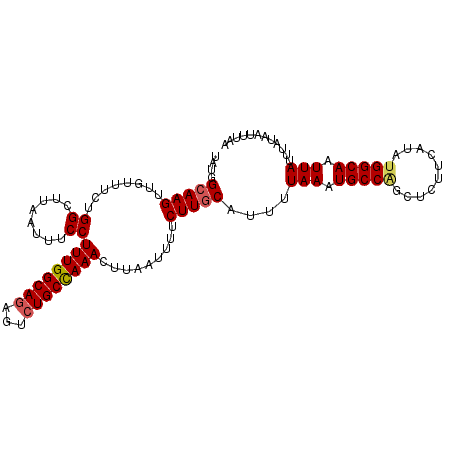

| Location | 26,993,242 – 26,993,358 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 85.53 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -21.22 |
| Energy contribution | -21.66 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26993242 116 - 27905053 UGCUGCUGCGAUCCAAUUGACCUUUUAGAGUGUAAAUAAAUAUGGAAAGUUGUUUCGGGCUUAAUUUCCUUUGGCAGAGUCUGCUAAACUUAAUUUUCUUCCAUUUUAAAUGCCAU .((.(((.(((..(((((..((.((((.........))))...))..)))))..)))))).........((((((((...)))))))).......................))... ( -20.10) >DroSec_CAF1 40745 107 - 1 ---------CAUCCAAUUGGCUUUUUGGAAUGUAAAUAAAUAUGGCAAGUUGUUUCUGGCUUAAUUUCCUUUGGCAGAGUCUGCUAAACUUAAUUUUCUUGCAUUUUAAAUGCCAG ---------........((((..((((((((((((..((((..((.((((((.........))))))))((((((((...))))))))....))))..)))))))))))).)))). ( -26.90) >DroSim_CAF1 40809 107 - 1 ---------CAUCCAAUUGGCUUUUUGGAAUGUAAAUAAAUAUGGCAAGUUGUUUCUGGCUUAAUUUCCUUUGGCAGAGUCUGCCAAACUUAAUUUUCUUGCAUUUUAAAUGCCUG ---------.........(((..((((((((((((..((((..((.((((((.........))))))))((((((((...))))))))....))))..)))))))))))).))).. ( -27.80) >DroEre_CAF1 43030 107 - 1 ---------UAUUUAAUUGGAAUCUUGGAGUGUAAAUAAAUAUGGCAAGCAGUUGCUGGCUCAAUUUCCUUUGGCAGAGUCUGCCAAACUUAAUUUUCUUGCAUUUUAAAUGCCAG ---------........(((....(((((((((((..((((.((((.(((....))).)).))......((((((((...))))))))....))))..)))))))))))...))). ( -28.20) >DroYak_CAF1 41487 107 - 1 ---------CAUCUAAUUGGUCUUUUGAAGUGUACAUAAAUAUGGCAAGUAGUUUUUGGCCAAAUAUCCUUUGGCAAAGUCUGCCAAACUUAAUUUUCUUGCAUUUUAAAUGCCGG ---------........((((..(((((((((((...((((.(((((((.....))).)))).......(((((((.....)))))))....))))...))))))))))).)))). ( -26.00) >consensus _________CAUCCAAUUGGCCUUUUGGAGUGUAAAUAAAUAUGGCAAGUUGUUUCUGGCUUAAUUUCCUUUGGCAGAGUCUGCCAAACUUAAUUUUCUUGCAUUUUAAAUGCCAG .................((((..((((((((((((..((((..(((.((......)).)))........((((((((...))))))))....))))..)))))))))))).)))). (-21.22 = -21.66 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:48:23 2006