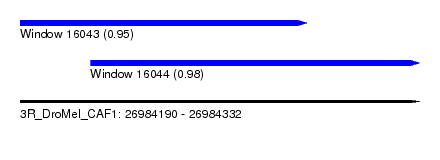
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,984,190 – 26,984,332 |
| Length | 142 |
| Max. P | 0.980256 |

| Location | 26,984,190 – 26,984,292 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 75.56 |
| Mean single sequence MFE | -48.05 |
| Consensus MFE | -22.77 |
| Energy contribution | -22.08 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26984190 102 + 27905053 GGAUCGAAAUCGGGUGGACAGGCCGCCACGUAGGCCUCUACCGCAGCCUUUUUGGCCUCCUCCGUAUACGCUCCAUCGCCGGUGGCCAUGAAAGCCGCCAAU (((.((....(((((((..(((((........)))))...)))).(((.....))).....)))....)).)))......((((((.......))))))... ( -39.30) >DroVir_CAF1 41728 95 + 1 GGC-------CGGCUUCGUUGGCCGCCACAUAGGCGGCAACGGCCGCUUGUUUGGCCUCGUCGGUGAUGGCGCCAUCGCCAGUGGCCAUAAAGGCGGCGACA (((-------((((...)))))))((((.(((((((((....))))))))).))))...((((((((((....))))))).((.(((.....))).))))). ( -56.10) >DroPse_CAF1 40064 102 + 1 GGAUCGGCUUCGGGUCUGCUGGCUGCCACAUAGGCGGCCACAGCCGCCUGCUUGGCCUCGGCAGUGAUUGCCCCAUCGCUGGCGGCCACAAAGGCGGCUGCC (((((.......)))))..((((((((.....))))))))(((((((((...(((((..(.(((((((......))))))).))))))...))))))))).. ( -56.70) >DroEre_CAF1 33917 102 + 1 GGAUCGAAAUCGGGCGGACAGGCUGCCACGUAGGCAUCUACCGCCGCCUGCUUCGCCUCCUCCGUAUACGCUCCAUCGCCGGUGGCCAUGAAAGCCGCCAAU (((.((....(((((((((((((.((...((((....)))).)).))))).))))).....)))....)).)))......((((((.......))))))... ( -39.20) >DroYak_CAF1 32294 102 + 1 GGAUCGAAAUCGGGCGGGCAGGCUGCCACGUAGGCCUCCACCGCCGCCUGUUUGGCCUCCUCCGUGUACGCUCCAUCGCCGGUGGCCAUGAAAGCCGCCAAU (((.((....((((((((.(((((........))))).).)))))(((.....)))......))....)).)))......((((((.......))))))... ( -40.30) >DroPer_CAF1 32639 102 + 1 GGAUCGGCUUCGGGUCUGCUGGCUGCCACAUAGGCGGCCACAGCCGCCUGCUUGGCCUCGGCAGUGAUUGCCCCAUCGCUGGCGGCCACAAAGGCGGCUGCC (((((.......)))))..((((((((.....))))))))(((((((((...(((((..(.(((((((......))))))).))))))...))))))))).. ( -56.70) >consensus GGAUCGAAAUCGGGUGGGCAGGCUGCCACAUAGGCGGCCACCGCCGCCUGCUUGGCCUCCUCCGUGAACGCUCCAUCGCCGGUGGCCAUAAAAGCCGCCAAU ............((((....((.((((.....)))).))...((((((.((.(((.................)))..)).)))))).........))))... (-22.77 = -22.08 + -0.69)


| Location | 26,984,215 – 26,984,332 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -53.18 |
| Consensus MFE | -26.63 |
| Energy contribution | -28.63 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26984215 117 + 27905053 CCACGUAGGCCUCUACCGCAGCCUUUUUGGCCUCCUCCGUAUACGCUCCAUCGCC---GGUGGCCAUGAAAGCCGCCAAUGCGGCCUUUUCCGGAUCCGUGACAGCCUGCAGGAGCUGCG ....((((....))))(((((((.....))).............(((((......---((((((.......))))))..(((((.((.(..((....))..).)).)))))))))))))) ( -41.60) >DroVir_CAF1 41746 117 + 1 CCACAUAGGCGGCAACGGCCGCUUGUUUGGCCUCGUCGGUGAUGGCGCCAUCGCC---AGUGGCCAUAAAGGCGGCGACAGCUGCCUUCUCAGCAUCGGUUACUGCCUCCAGAAGCUGCA ......(((((((....(((((((...(((((.(...(((((((....)))))))---.).)))))...)))))))....)))))))...((((.(((((....)))....)).)))).. ( -54.50) >DroGri_CAF1 42106 117 + 1 CCACAUAGGCGGCAAUAGCCGCCUGUUUGGCCUCCUCGGUGAUGACGCCAUCGCU---GGUGGCCACAAAGGCAGCGACAGCCGCCUUCUCCGCAUCGGUGACCGGCUCGAGCAGCUGCA ....(((((((((....))))))))).(((((.((..(((((((....)))))))---)).))))).....(((((..((((((...((.(((...))).)).)))))...)..))))). ( -56.10) >DroMoj_CAF1 43530 120 + 1 CAGCAUAGGCCUCAAUGGCCGCCUGCUUGGCCUCGCCCAUGUUGACGCCUUCGCCUGCGGUGGCCACAAAGGCGGCGACAGCUGCCUUCUCCGCAUCGGUGACAGCCUCCAGCAGCUGCA ..(((..((((.....))))((.((((.(((.(((((.((((.((.(((.(((....))).)))....((((((((....)))))))).)).)))).)))))..)))...))))))))). ( -54.60) >DroAna_CAF1 33945 117 + 1 GCGGGCAGGCGGCCACCGCUGCCUGCUUGGCCUCCGGCGUAAUUGCUCCAUCACU---GGUGGCCACGAAUGCCGCAACGGCAGCUUUUUCCGCAUCCGUUACAGCCUCAAGGAGCUGCA (((((((((((((....))))))))).(((((.((((.(............).))---)).)))))......))))....((((((((((..((..........))...)))))))))). ( -53.50) >DroPer_CAF1 32664 117 + 1 CCACAUAGGCGGCCACAGCCGCCUGCUUGGCCUCGGCAGUGAUUGCCCCAUCGCU---GGCGGCCACAAAGGCGGCUGCCGCGGCCUUUUCCGCAUCCGUGACUGCCUCCAGCAGCUGGA .......(((((.(((((((((((...(((((..(.(((((((......))))))---).))))))...))))))))...((((......))))....))).)))))(((((...))))) ( -58.80) >consensus CCACAUAGGCGGCAACAGCCGCCUGCUUGGCCUCCUCCGUGAUGGCGCCAUCGCC___GGUGGCCACAAAGGCGGCGACAGCGGCCUUCUCCGCAUCCGUGACAGCCUCCAGCAGCUGCA ....(((((((((....))))))))).(((((.(....(((((......))))).....).)))))..(((((.((....)).)))))................................ (-26.63 = -28.63 + 2.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:48:19 2006