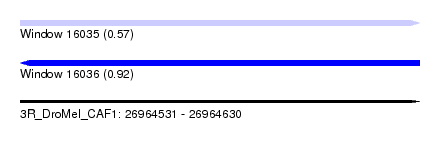
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,964,531 – 26,964,630 |
| Length | 99 |
| Max. P | 0.922233 |

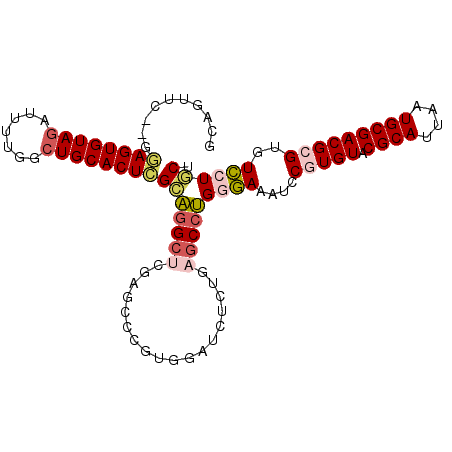
| Location | 26,964,531 – 26,964,630 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.41 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -17.75 |
| Energy contribution | -19.33 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26964531 99 + 27905053 AGCAGGACACGCGUCGCAUUAAUGCGUACACGGAUUUCGCAGGCUCAGAGAUCCACGGGCUCGAGCCUGCGAGUGCAGCCAGAAUCUACACUC----AAAUAC ....(((..((.((((((....)))).)).))...)))((((((((.(((.((....)))))))))))))(((((.((.......)).)))))----...... ( -36.80) >DroSec_CAF1 12082 102 + 1 AGCAGGACACGCGUCGCAUUAAUGCGUACAUGGAUUUCACAGGCUCAGAGAUCC-CAGGCUCCAGCGUGCGAGUGCAGCCAAAAUCUACACUCCCCGAACUGC .(((((.(((((((((((....)))).))..(((((((.........)))))))-.........))))))(((((.((.......)).)))))......)))) ( -31.60) >DroSim_CAF1 12375 103 + 1 AGCAGGACACGCGUCGCAUUAAUGCGUACACGGAUUUCCCAGGCUCAGAGAUCCACGGGCUCCAGCCUGCGAGUGCAGCCAAUCUCUACACUCCUCGAACUGC .((((..(..((((((((....)))).))..(((((((.........)))))))..((((....))))))(((((.((.......)).)))))...)..)))) ( -31.80) >DroEre_CAF1 14134 76 + 1 AGCAGGACACGCGUCGCAUUAAUGCGAACAGGGAUUUCCCAGGC----------------UCGAGCCUGCGAGUGCAGCCAAAAUCUACACU----------- .(((((.(.((.((((((....))).....(((....))).)))----------------.)).)))))).((((.((.......)).))))----------- ( -24.80) >DroYak_CAF1 12136 103 + 1 AGCAGGACACGCGUCGCAUUAAUGCGUACACGGAUUUCCCAGGCUCGCAGAUCCACGGGCUCGAGCCUCCGAGUGCAGCCAAAAUCUACACUCCUCGAACUGC .((((..(.((.((((((....)))).)).))((......(((((((.((.((....)))))))))))..(((((.((.......)).))))).)))..)))) ( -32.70) >DroAna_CAF1 12881 101 + 1 AGUAGAACACGCGUCGCAUUAAUGCGUACACGGAUUUUCCUGGCUAGGAAAUUCAGUAGGCUGGCCCAACAAGUGCAGCCUAAAGAUACACUUG--GAAAAAU ((.((((..((.((((((....)))).)).))..)))).))..(((((....((..(((((((((.......)).)))))))..))....))))--)...... ( -26.30) >consensus AGCAGGACACGCGUCGCAUUAAUGCGUACACGGAUUUCCCAGGCUCAGAGAUCCACGGGCUCGAGCCUGCGAGUGCAGCCAAAAUCUACACUCC__GAACUGC .(((((......((((((....)))).))..(((((((.........)))))))...........)))))(((((.((.......)).))))).......... (-17.75 = -19.33 + 1.59)

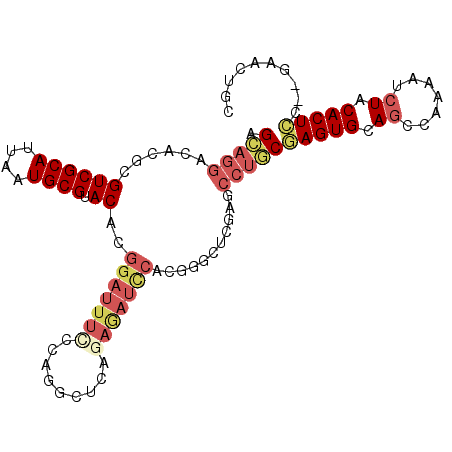

| Location | 26,964,531 – 26,964,630 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.41 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -23.62 |
| Energy contribution | -24.45 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26964531 99 - 27905053 GUAUUU----GAGUGUAGAUUCUGGCUGCACUCGCAGGCUCGAGCCCGUGGAUCUCUGAGCCUGCGAAAUCCGUGUACGCAUUAAUGCGACGCGUGUCCUGCU ......----((((((((.......))))))))(((((((((((.(....)..))).))))))))......(((((.((((....)))))))))......... ( -41.10) >DroSec_CAF1 12082 102 - 1 GCAGUUCGGGGAGUGUAGAUUUUGGCUGCACUCGCACGCUGGAGCCUG-GGAUCUCUGAGCCUGUGAAAUCCAUGUACGCAUUAAUGCGACGCGUGUCCUGCU ((((......((((((((.......))))))))((((((((((.(.((-((.((...)).)))).)...)))).((.((((....)))))))))))).)))). ( -37.50) >DroSim_CAF1 12375 103 - 1 GCAGUUCGAGGAGUGUAGAGAUUGGCUGCACUCGCAGGCUGGAGCCCGUGGAUCUCUGAGCCUGGGAAAUCCGUGUACGCAUUAAUGCGACGCGUGUCCUGCU ((((..((.(((((((((.......))))))((.((((((((((.(....)..)))).)))))).))..)))((((.((((....)))))))).))..)))). ( -42.80) >DroEre_CAF1 14134 76 - 1 -----------AGUGUAGAUUUUGGCUGCACUCGCAGGCUCGA----------------GCCUGGGAAAUCCCUGUUCGCAUUAAUGCGACGCGUGUCCUGCU -----------(((((((.......))))))).((((((.(((----------------((..(((....))).))))(((....))).....).).))))). ( -28.20) >DroYak_CAF1 12136 103 - 1 GCAGUUCGAGGAGUGUAGAUUUUGGCUGCACUCGGAGGCUCGAGCCCGUGGAUCUGCGAGCCUGGGAAAUCCGUGUACGCAUUAAUGCGACGCGUGUCCUGCU ((((..((..((((((((.......))))))))(((..((((.((.(((......))).)).))))...)))((((.((((....)))))))).))..)))). ( -41.30) >DroAna_CAF1 12881 101 - 1 AUUUUUC--CAAGUGUAUCUUUAGGCUGCACUUGUUGGGCCAGCCUACUGAAUUUCCUAGCCAGGAAAAUCCGUGUACGCAUUAAUGCGACGCGUGUUCUACU .......--..((((..((..(((((((..((....))..)))))))..)).((((((....))))))...(((((.((((....))))))))).....)))) ( -29.30) >consensus GCAGUUC__GGAGUGUAGAUUUUGGCUGCACUCGCAGGCUCGAGCCCGUGGAUCUCUGAGCCUGGGAAAUCCGUGUACGCAUUAAUGCGACGCGUGUCCUGCU ..........((((((((.......))))))))(((((((..................)))))((((....(((((.((((....)))))))))..)))))). (-23.62 = -24.45 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:48:12 2006