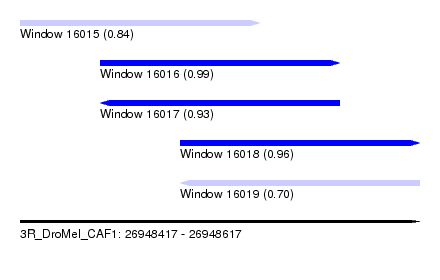
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,948,417 – 26,948,617 |
| Length | 200 |
| Max. P | 0.989775 |

| Location | 26,948,417 – 26,948,537 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -30.02 |
| Energy contribution | -29.38 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26948417 120 + 27905053 AUAUGAAAGACGGUGGUGUUACCGUAACGCUGUACGAAAGCAAUUUCCAGUUUUUCGGGGGUCAAGCUAUCUCUGCCCAAGGUCAUGCCCAUGACCUUGAUGGCUACAUCCUUGGCCUCC ....(((((((.(..((((((...))))))..)..((((....))))..)))))))((((((((((........((((((((((((....)))))))))..)))......)))))))))) ( -47.54) >DroSec_CAF1 16083 120 + 1 AUAUGAAAGACAGUGGUGUCACAGAAACGCUGUAUGGAAGCAAUUUCCAGGUUUGCGGGGGUCAAGCUACCUCUGUCCAAGGUCAUGCCCAUGACCUUGAUGGCUACGUCCUUGGCUUCC .........((((((...((...))..))))))..((((((........((...((((((((......))))))))))((((((((....))))))))((((....))))....)))))) ( -39.80) >DroSim_CAF1 16138 120 + 1 AUAUGAAAGACAGUGGUGCCACAGAAACGCUGUAUGGAAGCAAUUUCCAGGUUUGCGGGGGUCAAGCUACCUCUGUCCAAGGUCAUGCCCAUGACCUUGAUGGCUACGUCCUUGGCUUCC .........((((((............))))))..((((((........((...((((((((......))))))))))((((((((....))))))))((((....))))....)))))) ( -39.60) >DroEre_CAF1 18758 120 + 1 AUAUGAAAUAUGGUGGUGUUAUAGAAGCGCUGUAGUACAGCCAUUUCCAGCUUCGCAGGGGUCAAGCUAUCUCUAUCCAAGGUCAUGCCCAUGACCUUGAUGGCUACAUCCUUGGCUUCC ...........((.(((......(((((((((.....))))........))))).(((((((..(((((((.......((((((((....)))))))))))))))..)))))))))).)) ( -38.51) >DroYak_CAF1 16583 120 + 1 GUUUGAAAGACGCUGGUGUUAUAGAAGCGCUGUACUAGAGCCAUUUCCAGCUUGGCAGGAUUCAAGCUAUCUGUACCCAAGGUCAUGCCCAUGACCUUGAUGGCCACAUCCUUGGCCUCC (((((((...((((...........))))((((....((((........)))).))))..)))))))..........(((((((((....)))))))))..(((((......)))))... ( -39.40) >consensus AUAUGAAAGACGGUGGUGUUACAGAAACGCUGUACGAAAGCAAUUUCCAGCUUUGCGGGGGUCAAGCUAUCUCUGUCCAAGGUCAUGCCCAUGACCUUGAUGGCUACAUCCUUGGCUUCC ........(((((.(((...((((.....)))).....(((...((((.(.....).))))....)))))).)))))(((((((((....)))))))))..(((((......)))))... (-30.02 = -29.38 + -0.64)


| Location | 26,948,457 – 26,948,577 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -42.67 |
| Consensus MFE | -33.07 |
| Energy contribution | -34.35 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26948457 120 + 27905053 CAAUUUCCAGUUUUUCGGGGGUCAAGCUAUCUCUGCCCAAGGUCAUGCCCAUGACCUUGAUGGCUACAUCCUUGGCCUCCUCGACCGAAGGACUUACGUAGCCCUUGCUAAAUAGUUCCU ................((((((((((........((((((((((((....)))))))))..)))......))))))))))........((((.(((..((((....))))..))).)))) ( -41.84) >DroSec_CAF1 16123 120 + 1 CAAUUUCCAGGUUUGCGGGGGUCAAGCUACCUCUGUCCAAGGUCAUGCCCAUGACCUUGAUGGCUACGUCCUUGGCUUCCUCGAGCGAAGGACAUACGUAACCCUUGCUGAAUAGUUCAU ..........((((((((((((..(((((........(((((((((....))))))))).)))))..((((((.((((....)))).)))))).......)))))))).))))....... ( -43.30) >DroSim_CAF1 16178 120 + 1 CAAUUUCCAGGUUUGCGGGGGUCAAGCUACCUCUGUCCAAGGUCAUGCCCAUGACCUUGAUGGCUACGUCCUUGGCUUCCUCGAGCGAAGGACAUACGUAACCCUUGCUGAAUAGUUCAU ..........((((((((((((..(((((........(((((((((....))))))))).)))))..((((((.((((....)))).)))))).......)))))))).))))....... ( -43.30) >DroEre_CAF1 18798 120 + 1 CCAUUUCCAGCUUCGCAGGGGUCAAGCUAUCUCUAUCCAAGGUCAUGCCCAUGACCUUGAUGGCUACAUCCUUGGCUUCCUCGAGCGAGGGACUUGCGUAGCCCUUGCUGAAUAGUUCCU .........(((.((.((((((((((......(((((.((((((((....))))))))))))).......)))))).))))))))).(((((((....((((....))))...))))))) ( -44.42) >DroYak_CAF1 16623 120 + 1 CCAUUUCCAGCUUGGCAGGAUUCAAGCUAUCUGUACCCAAGGUCAUGCCCAUGACCUUGAUGGCCACAUCCUUGGCCUCCUCGAGCGAGGGACACACGUAGCCCUUGGUGAAGAGUUCCU ........(((((((......))))))).(((.(((((((((((((....)))))))))..(((((......)))))(((((....)))))...............)))).)))...... ( -40.50) >consensus CAAUUUCCAGCUUUGCGGGGGUCAAGCUAUCUCUGUCCAAGGUCAUGCCCAUGACCUUGAUGGCUACAUCCUUGGCUUCCUCGAGCGAAGGACAUACGUAGCCCUUGCUGAAUAGUUCCU ..............((((((((..((((((.......(((((((((....)))))))))))))))...(((((.((((....)))).)))))........))))))))............ (-33.07 = -34.35 + 1.28)


| Location | 26,948,457 – 26,948,577 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -42.06 |
| Consensus MFE | -34.06 |
| Energy contribution | -33.82 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26948457 120 - 27905053 AGGAACUAUUUAGCAAGGGCUACGUAAGUCCUUCGGUCGAGGAGGCCAAGGAUGUAGCCAUCAAGGUCAUGGGCAUGACCUUGGGCAGAGAUAGCUUGACCCCCGAAAAACUGGAAAUUG .....((((((.((...((((((....((((((.((((.....)))))))))))))))).((((((((((....))))))))))))..))))))........(((......)))...... ( -42.70) >DroSec_CAF1 16123 120 - 1 AUGAACUAUUCAGCAAGGGUUACGUAUGUCCUUCGCUCGAGGAAGCCAAGGACGUAGCCAUCAAGGUCAUGGGCAUGACCUUGGACAGAGGUAGCUUGACCCCCGCAAACCUGGAAAUUG ............((..((((((.((((((((((.(((......))).)))))))).(((.((((((((((....)))))))))).....))).)).))))))..)).............. ( -40.40) >DroSim_CAF1 16178 120 - 1 AUGAACUAUUCAGCAAGGGUUACGUAUGUCCUUCGCUCGAGGAAGCCAAGGACGUAGCCAUCAAGGUCAUGGGCAUGACCUUGGACAGAGGUAGCUUGACCCCCGCAAACCUGGAAAUUG ............((..((((((.((((((((((.(((......))).)))))))).(((.((((((((((....)))))))))).....))).)).))))))..)).............. ( -40.40) >DroEre_CAF1 18798 120 - 1 AGGAACUAUUCAGCAAGGGCUACGCAAGUCCCUCGCUCGAGGAAGCCAAGGAUGUAGCCAUCAAGGUCAUGGGCAUGACCUUGGAUAGAGAUAGCUUGACCCCUGCGAAGCUGGAAAUGG ........((((((..(((..((....)))))((((..(.((((((...((......)).((((((((((....)))))))))).........))))..)))..)))).))))))..... ( -40.10) >DroYak_CAF1 16623 120 - 1 AGGAACUCUUCACCAAGGGCUACGUGUGUCCCUCGCUCGAGGAGGCCAAGGAUGUGGCCAUCAAGGUCAUGGGCAUGACCUUGGGUACAGAUAGCUUGAAUCCUGCCAAGCUGGAAAUGG ......(((..(((...((((((((.((((((((....)))).)))...).))))))))..(((((((((....))))))))))))..)))(((((((........)))))))....... ( -46.70) >consensus AGGAACUAUUCAGCAAGGGCUACGUAUGUCCUUCGCUCGAGGAAGCCAAGGAUGUAGCCAUCAAGGUCAUGGGCAUGACCUUGGACAGAGAUAGCUUGACCCCCGCAAACCUGGAAAUUG ........(((((((((((((((....((((((.(((......))).)))))))))))).((((((((((....))))))))))..........))))............)))))..... (-34.06 = -33.82 + -0.24)

| Location | 26,948,497 – 26,948,617 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -41.54 |
| Consensus MFE | -33.34 |
| Energy contribution | -34.86 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26948497 120 + 27905053 GGUCAUGCCCAUGACCUUGAUGGCUACAUCCUUGGCCUCCUCGACCGAAGGACUUACGUAGCCCUUGCUAAAUAGUUCCUUCUGAAGCAUUUCCAUGGCUGCCCCUGACUUGCGGCCAAU ((((((....))))))..((((....)))).((((((.........((((((.(((..((((....))))..))).))))))....(((..((...((.....)).))..))))))))). ( -39.50) >DroSec_CAF1 16163 120 + 1 GGUCAUGCCCAUGACCUUGAUGGCUACGUCCUUGGCUUCCUCGAGCGAAGGACAUACGUAACCCUUGCUGAAUAGUUCAUUCUGAAGCAUUUCCAUGGCUGCCCCUGACUUGCGGCCAAU ((((((....))))))...((((....((((((.((((....)))).))))))............((((...(((......))).))))...))))((((((.........))))))... ( -39.90) >DroSim_CAF1 16218 120 + 1 GGUCAUGCCCAUGACCUUGAUGGCUACGUCCUUGGCUUCCUCGAGCGAAGGACAUACGUAACCCUUGCUGAAUAGUUCAUUCUGAAGCAUUUCCAAGGCUGCCCCUGACUUGCGGCCAAU ((((((....))))))....(((....((((((.((((....)))).))))))............((((...(((......))).))))...))).((((((.........))))))... ( -39.00) >DroEre_CAF1 18838 120 + 1 GGUCAUGCCCAUGACCUUGAUGGCUACAUCCUUGGCUUCCUCGAGCGAGGGACUUGCGUAGCCCUUGCUGAAUAGUUCCUUCUGAAGCAUUUCCAUGGCCGCCCCUGAUUUGCGGCCAAU ((((((....))))))..((((....))))....(((((.((.((((((((.((.....))))))))))))..((......))))))).......(((((((.........))))))).. ( -44.20) >DroYak_CAF1 16663 120 + 1 GGUCAUGCCCAUGACCUUGAUGGCCACAUCCUUGGCCUCCUCGAGCGAGGGACACACGUAGCCCUUGGUGAAGAGUUCCUUCUGGAGCAGUUCCAUGGCCGCCCCUGAAUGGCGACCAAU ((((..(.(((((..(((((.(((((......)))))...)))))((((((.(.......)))))))..(((..(((((....)))))..)))))))).)(((.......)))))))... ( -45.10) >consensus GGUCAUGCCCAUGACCUUGAUGGCUACAUCCUUGGCUUCCUCGAGCGAAGGACAUACGUAGCCCUUGCUGAAUAGUUCCUUCUGAAGCAUUUCCAUGGCUGCCCCUGACUUGCGGCCAAU ((((((....)))))).....((((((.(((((.((((....)))).))))).....))))))..((((...(((......))).))))......(((((((.........))))))).. (-33.34 = -34.86 + 1.52)


| Location | 26,948,497 – 26,948,617 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -42.46 |
| Consensus MFE | -34.00 |
| Energy contribution | -33.60 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26948497 120 - 27905053 AUUGGCCGCAAGUCAGGGGCAGCCAUGGAAAUGCUUCAGAAGGAACUAUUUAGCAAGGGCUACGUAAGUCCUUCGGUCGAGGAGGCCAAGGAUGUAGCCAUCAAGGUCAUGGGCAUGACC ..((((..((.....((((((.(....)...)))))).((((((..(((.((((....)))).)))..))))))((((.....)))).....))..))))....((((((....)))))) ( -44.30) >DroSec_CAF1 16163 120 - 1 AUUGGCCGCAAGUCAGGGGCAGCCAUGGAAAUGCUUCAGAAUGAACUAUUCAGCAAGGGUUACGUAUGUCCUUCGCUCGAGGAAGCCAAGGACGUAGCCAUCAAGGUCAUGGGCAUGACC ..((((.((......((((((.(....)...)))))).(((((...))))).))..........(((((((((.(((......))).)))))))))))))....((((((....)))))) ( -38.50) >DroSim_CAF1 16218 120 - 1 AUUGGCCGCAAGUCAGGGGCAGCCUUGGAAAUGCUUCAGAAUGAACUAUUCAGCAAGGGUUACGUAUGUCCUUCGCUCGAGGAAGCCAAGGACGUAGCCAUCAAGGUCAUGGGCAUGACC ..((((.((......((((((.(....)...)))))).(((((...))))).))..........(((((((((.(((......))).)))))))))))))....((((((....)))))) ( -38.50) >DroEre_CAF1 18838 120 - 1 AUUGGCCGCAAAUCAGGGGCGGCCAUGGAAAUGCUUCAGAAGGAACUAUUCAGCAAGGGCUACGCAAGUCCCUCGCUCGAGGAAGCCAAGGAUGUAGCCAUCAAGGUCAUGGGCAUGACC ..(((((((.........)))))))(((...(((.((....((.....((((((.((((..((....)))))).))).)))....))...)).))).)))....((((((....)))))) ( -42.70) >DroYak_CAF1 16663 120 - 1 AUUGGUCGCCAUUCAGGGGCGGCCAUGGAACUGCUCCAGAAGGAACUCUUCACCAAGGGCUACGUGUGUCCCUCGCUCGAGGAGGCCAAGGAUGUGGCCAUCAAGGUCAUGGGCAUGACC ..((((((((.......))))))))((((.....))))((((.....))))......((((((((.((((((((....)))).)))...).)))))))).....((((((....)))))) ( -48.30) >consensus AUUGGCCGCAAGUCAGGGGCAGCCAUGGAAAUGCUUCAGAAGGAACUAUUCAGCAAGGGCUACGUAUGUCCUUCGCUCGAGGAAGCCAAGGAUGUAGCCAUCAAGGUCAUGGGCAUGACC .((((..((......((((((.(....)...)))))).((((.....)))).))...((((((....((((((.(((......))).)))))))))))).))))((((((....)))))) (-34.00 = -33.60 + -0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:47:56 2006