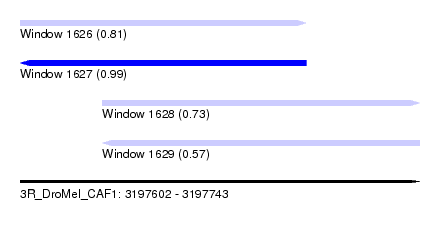
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,197,602 – 3,197,743 |
| Length | 141 |
| Max. P | 0.994217 |

| Location | 3,197,602 – 3,197,703 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.07 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -16.68 |
| Energy contribution | -18.06 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3197602 101 + 27905053 GGUG-------UAGAUACAAUAUGGGCCUUCUUCUC-----G-------GUUGCGUAUACGCAACAUAUGCGAGAAUGAAAAAUGAAAAUAAUUGUUGAAAAACUGAUAGCUAUUCGGCU ....-------.............((((((((((((-----(-------((((((....)))))).....)))))).)))..............(((....)))............)))) ( -22.70) >DroSec_CAF1 17172 108 + 1 GGUG-------UAGAUACAAUAUUUGCCUUCUUCUC-----GGGCACGCGUUGCGUAUACGCAACGUAUGCGAGAAUGAAAAAUGAAAAUAAUUGUUGAAAACCUGCUAGCCAUUCGGCU (((.-------.....(((((((((...((((((((-----..(((..(((((((....)))))))..)))))))).)))......)))).))))).....)))....((((....)))) ( -34.00) >DroSim_CAF1 23984 108 + 1 GGUG-------UAGAUACAAUAUGUGCCUUCUUCUC-----GGGCACGCGUUGCGUAUACGCAACGUAUGCGAGAAUGAAAAAUGAAAAUAAUUGUUGAAAACCUGCUAGCCAUUCGGCU (((.-------.....(((((.......((((((((-----..(((..(((((((....)))))))..)))))))).)))..((....)).))))).....)))....((((....)))) ( -33.00) >DroEre_CAF1 16777 109 + 1 UGUGCCUACAAUAUAUACCAUAUGUGCCUCCAACUCUGUUGGGCCACACGUUGCGUAUACGCAACGUAUGCGGGAAUGAAAA-----------UUUUUAAAAACUGCCCGCUAUGCGGCU ...(((................((((.(.(((((...)))))).)))).((((((....))))))(((((((((........-----------.............))))).))))))). ( -33.60) >consensus GGUG_______UAGAUACAAUAUGUGCCUUCUUCUC_____GGGCACGCGUUGCGUAUACGCAACGUAUGCGAGAAUGAAAAAUGAAAAUAAUUGUUGAAAAACUGCUAGCCAUUCGGCU .........................(((...............(((.((((((((....)))))))).)))..(((((.................................)))))))). (-16.68 = -18.06 + 1.38)

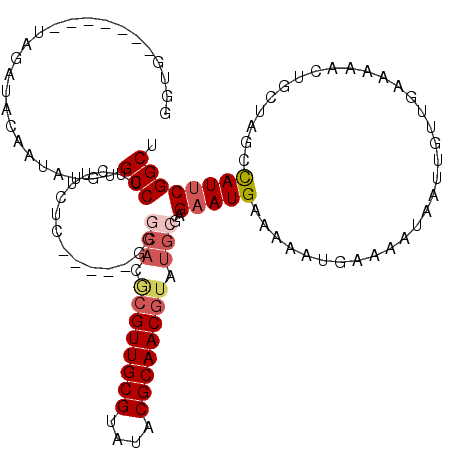
| Location | 3,197,602 – 3,197,703 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.07 |
| Mean single sequence MFE | -31.09 |
| Consensus MFE | -17.52 |
| Energy contribution | -20.34 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3197602 101 - 27905053 AGCCGAAUAGCUAUCAGUUUUUCAACAAUUAUUUUCAUUUUUCAUUCUCGCAUAUGUUGCGUAUACGCAAC-------C-----GAGAAGAAGGCCCAUAUUGUAUCUA-------CACC .((((((.(((.....))).))).................(((.((((((.....((((((....))))))-------)-----)))))))))))..............-------.... ( -23.60) >DroSec_CAF1 17172 108 - 1 AGCCGAAUGGCUAGCAGGUUUUCAACAAUUAUUUUCAUUUUUCAUUCUCGCAUACGUUGCGUAUACGCAACGCGUGCCC-----GAGAAGAAGGCAAAUAUUGUAUCUA-------CACC ((((....))))....(((.....(((((.((((....(((((.((((((((..(((((((....)))))))..)))..-----)))))))))).))))))))).....-------.))) ( -31.10) >DroSim_CAF1 23984 108 - 1 AGCCGAAUGGCUAGCAGGUUUUCAACAAUUAUUUUCAUUUUUCAUUCUCGCAUACGUUGCGUAUACGCAACGCGUGCCC-----GAGAAGAAGGCACAUAUUGUAUCUA-------CACC .((((((.((((....))))))).................(((.((((((((..(((((((....)))))))..)))..-----)))))))))))..............-------.... ( -30.80) >DroEre_CAF1 16777 109 - 1 AGCCGCAUAGCGGGCAGUUUUUAAAAA-----------UUUUCAUUCCCGCAUACGUUGCGUAUACGCAACGUGUGGCCCAACAGAGUUGGAGGCACAUAUGGUAUAUAUUGUAGGCACA .((((((..(((((.(((.........-----------.....))))))))((((((((((....))))))(((((.((((((...))))).).)))))...))))....))).)))... ( -38.84) >consensus AGCCGAAUAGCUAGCAGGUUUUCAACAAUUAUUUUCAUUUUUCAUUCUCGCAUACGUUGCGUAUACGCAACGCGUGCCC_____GAGAAGAAGGCACAUAUUGUAUCUA_______CACC .((((((.(((.....))).))).................(((.(((((((((.(((((((....))))))).)))).......)))))))))))......................... (-17.52 = -20.34 + 2.81)

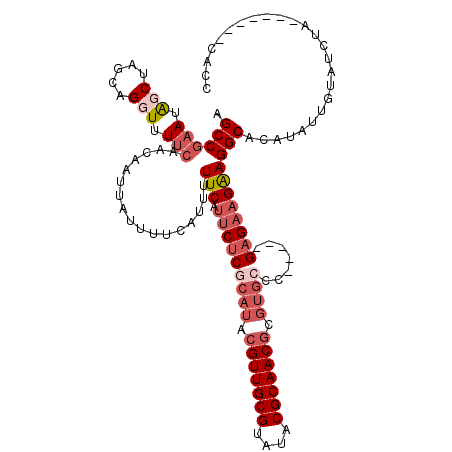
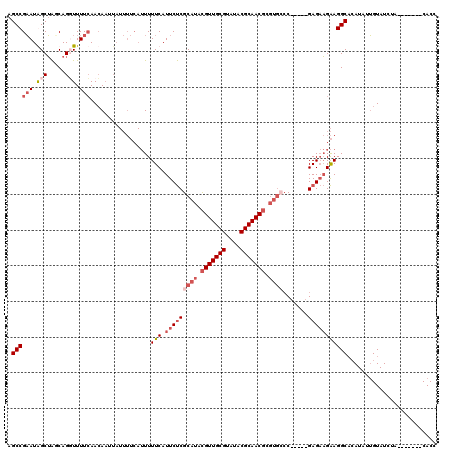
| Location | 3,197,631 – 3,197,743 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -33.21 |
| Consensus MFE | -22.84 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3197631 112 + 27905053 -G-------GUUGCGUAUACGCAACAUAUGCGAGAAUGAAAAAUGAAAAUAAUUGUUGAAAAACUGAUAGCUAUUCGGCUCACUCCUCUCAGCAGUGUUCGUCCCAUCCUAGCUCCCAUG -.-------((((((....)))))).((((.(((...((...(((((....((((((((.........((((....))))........)))))))).)))))....))....))).)))) ( -26.33) >DroSec_CAF1 17201 119 + 1 -GGGCACGCGUUGCGUAUACGCAACGUAUGCGAGAAUGAAAAAUGAAAAUAAUUGUUGAAAACCUGCUAGCCAUUCGGCUCACUCCUCUCAGGAGUGUUCGUCCUAUCCUAGCUCCCAUG -(((((..(((((((....)))))))..)))(((...((...(((((.....................((((....))))((((((.....)))))))))))....))....)))))... ( -37.90) >DroSim_CAF1 24013 119 + 1 -GGGCACGCGUUGCGUAUACGCAACGUAUGCGAGAAUGAAAAAUGAAAAUAAUUGUUGAAAACCUGCUAGCCAUUCGGCUCACUCCUCUCAGGAGUGUUCGUCCUAUCCUAGCUCCCAUG -(((((..(((((((....)))))))..)))(((...((...(((((.....................((((....))))((((((.....)))))))))))....))....)))))... ( -37.90) >DroEre_CAF1 16817 109 + 1 GGGCCACACGUUGCGUAUACGCAACGUAUGCGGGAAUGAAAA-----------UUUUUAAAAACUGCCCGCUAUGCGGCUCACUCCUCUCAGCAGUAUUCGUGUUAUCCUAGCUCCCAUC (((((....((((((....))))))(((((((((........-----------.............))))).))))))))).........(((((.((.......)).)).)))...... ( -30.70) >consensus _GGGCACGCGUUGCGUAUACGCAACGUAUGCGAGAAUGAAAAAUGAAAAUAAUUGUUGAAAAACUGCUAGCCAUUCGGCUCACUCCUCUCAGCAGUGUUCGUCCUAUCCUAGCUCCCAUG .......((((((((....))))))))(((.(((.(((((.............................(((....))).((((((.....)))))))))))..........))).))). (-22.84 = -23.52 + 0.69)

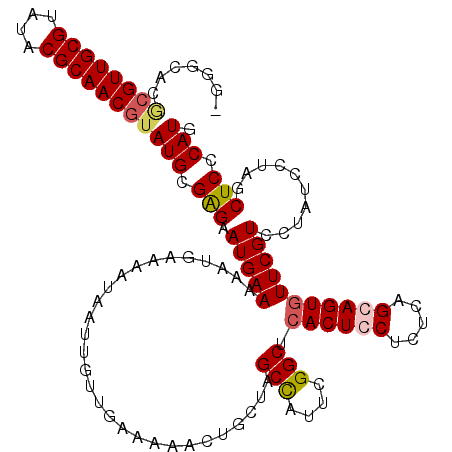
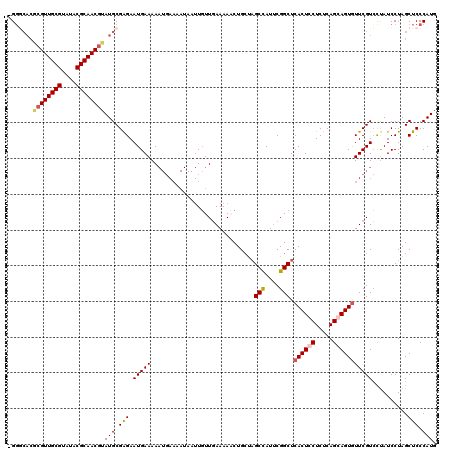
| Location | 3,197,631 – 3,197,743 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -23.77 |
| Energy contribution | -25.65 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3197631 112 - 27905053 CAUGGGAGCUAGGAUGGGACGAACACUGCUGAGAGGAGUGAGCCGAAUAGCUAUCAGUUUUUCAACAAUUAUUUUCAUUUUUCAUUCUCGCAUAUGUUGCGUAUACGCAAC-------C- .......((.((((((..(....((((.((....))))))....(((.(((.....))).)))................)..)))))).))....((((((....))))))-------.- ( -28.90) >DroSec_CAF1 17201 119 - 1 CAUGGGAGCUAGGAUAGGACGAACACUCCUGAGAGGAGUGAGCCGAAUGGCUAGCAGGUUUUCAACAAUUAUUUUCAUUUUUCAUUCUCGCAUACGUUGCGUAUACGCAACGCGUGCCC- ..((..((((.............((((((.....))))))((((....))))....))))..)).........................(((..(((((((....)))))))..)))..- ( -38.80) >DroSim_CAF1 24013 119 - 1 CAUGGGAGCUAGGAUAGGACGAACACUCCUGAGAGGAGUGAGCCGAAUGGCUAGCAGGUUUUCAACAAUUAUUUUCAUUUUUCAUUCUCGCAUACGUUGCGUAUACGCAACGCGUGCCC- ..((..((((.............((((((.....))))))((((....))))....))))..)).........................(((..(((((((....)))))))..)))..- ( -38.80) >DroEre_CAF1 16817 109 - 1 GAUGGGAGCUAGGAUAACACGAAUACUGCUGAGAGGAGUGAGCCGCAUAGCGGGCAGUUUUUAAAAA-----------UUUUCAUUCCCGCAUACGUUGCGUAUACGCAACGUGUGGCCC ...(((((..(((((.....(((.((((((....).......((((...))))))))).)))....)-----------))))..))))).(((((((((((....))))))))))).... ( -36.30) >consensus CAUGGGAGCUAGGAUAGGACGAACACUCCUGAGAGGAGUGAGCCGAAUAGCUAGCAGGUUUUCAACAAUUAUUUUCAUUUUUCAUUCUCGCAUACGUUGCGUAUACGCAACGCGUGCCC_ ...(((((..((((.........((((((.....))))))(((......)))..........................))))..)))))((((.(((((((....))))))).))))... (-23.77 = -25.65 + 1.88)

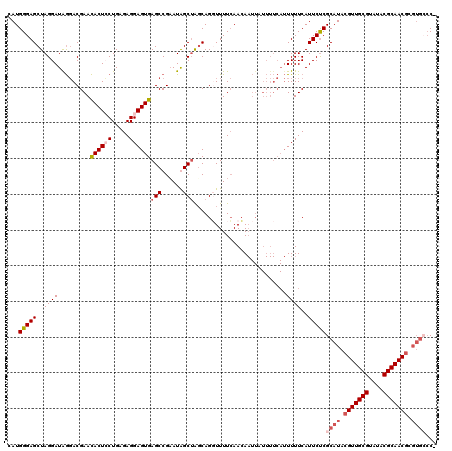
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:10 2006