
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,929,108 – 26,929,265 |
| Length | 157 |
| Max. P | 0.925921 |

| Location | 26,929,108 – 26,929,207 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -22.76 |
| Energy contribution | -24.93 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
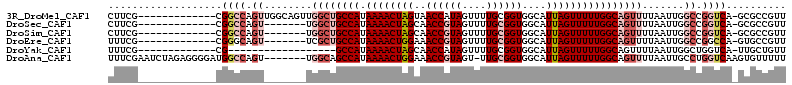
>3R_DroMel_CAF1 26929108 99 + 27905053 CUUCG-------------CGGCCAGUUGGCAGUUGGCUGCCAUAAAACUAGUAACCAUAGUUUUGCGGUGGCAUUAGUUUUUGGCAGUUUUAAUUGGCCGGUCA-GCGCCGUU .....-------------.((((((((((.....((((((((.(((((((((..((((.(.....).)))).)))))))))))))))))))))))))))(((..-..)))... ( -38.40) >DroSec_CAF1 3871 92 + 1 CUUCG-------------CGGCCAGU-------UGGCUGCCAUAAAACUAGCAACCGUAGUUUUGCGGUGGCAUUAGUUUUUGGCAGUUUUAAUUGGCCGGUCA-GCGCCGUU .....-------------.(((((((-------(((((((((.(((((((((.((((((....)))))).))..))))))))))))))..)))))))))(((..-..)))... ( -42.00) >DroSim_CAF1 3923 92 + 1 CUUCG-------------CGGCCAGU-------UGGCUGCCAUAAAACUAGCAACCGUAGUUUUGCGGUGGCAUUAGUUUUUGGCAGUUUUAAUUGGCCGGUCA-GCGCCGUU .....-------------.(((((((-------(((((((((.(((((((((.((((((....)))))).))..))))))))))))))..)))))))))(((..-..)))... ( -42.00) >DroEre_CAF1 4063 92 + 1 UUUCG-------------CGGGCAGU-------UCGCUGCCAUAAAACUGGAAACCGUAGUUUUGCGGUGGCAUUAGUUUUUGGCAGUUUUAAUUGGCCGGCCA-GUGCCGUU .....-------------.((.((((-------(.(((((((.((((((((..((((((....))))))....)))))))))))))))...))))).))(((..-..)))... ( -35.60) >DroYak_CAF1 4181 81 + 1 UUUCG-------------CG------------------GCCAUAAAACUAGCAACCAUAGUUUUGCGGUGGCAUUAGUUUUUGGCAGUUUUAAUUGGCUGGUCA-UUGCUGUU ....(-------------(.------------------(((.((((((((.......)))))))).))).))..((((...((((((((......)))).))))-..)))).. ( -24.10) >DroAna_CAF1 4262 105 + 1 UUUCGAAUCUAGAGGGGAUGGCCAGU-------UGGCAGCCAUAAAACUGGAAACCGUAGU-UUGCGGUGGCAUUAGUUUUUGGCAGUUUUAAUUGCCUGGUCAAGUGUUUUU ......((((.....))))(((((..-------.((((((((.((((((((..(((((...-..)))))....)))))))))))).........))))))))).......... ( -28.40) >consensus CUUCG_____________CGGCCAGU_______UGGCUGCCAUAAAACUAGCAACCGUAGUUUUGCGGUGGCAUUAGUUUUUGGCAGUUUUAAUUGGCCGGUCA_GCGCCGUU ...................((((.((........((((((((.((((((((..((((((....))))))....)))))))))))))))).......)).)))).......... (-22.76 = -24.93 + 2.17)

| Location | 26,929,108 – 26,929,207 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -15.58 |
| Energy contribution | -16.92 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26929108 99 - 27905053 AACGGCGC-UGACCGGCCAAUUAAAACUGCCAAAAACUAAUGCCACCGCAAAACUAUGGUUACUAGUUUUAUGGCAGCCAACUGCCAACUGGCCG-------------CGAAG ........-...(((((((..(((((((((((........(((....)))......))))....)))))))((((((....))))))..))))))-------------.)... ( -27.94) >DroSec_CAF1 3871 92 - 1 AACGGCGC-UGACCGGCCAAUUAAAACUGCCAAAAACUAAUGCCACCGCAAAACUACGGUUGCUAGUUUUAUGGCAGCCA-------ACUGGCCG-------------CGAAG ........-...(((((((.......(((((((((((((..((.((((........)))).))))))))).))))))...-------..))))))-------------.)... ( -32.00) >DroSim_CAF1 3923 92 - 1 AACGGCGC-UGACCGGCCAAUUAAAACUGCCAAAAACUAAUGCCACCGCAAAACUACGGUUGCUAGUUUUAUGGCAGCCA-------ACUGGCCG-------------CGAAG ........-...(((((((.......(((((((((((((..((.((((........)))).))))))))).))))))...-------..))))))-------------.)... ( -32.00) >DroEre_CAF1 4063 92 - 1 AACGGCAC-UGGCCGGCCAAUUAAAACUGCCAAAAACUAAUGCCACCGCAAAACUACGGUUUCCAGUUUUAUGGCAGCGA-------ACUGCCCG-------------CGAAA ..((((..-..))))((((..((((((((...........(((....)))((((....)))).)))))))))))).(((.-------......))-------------).... ( -27.20) >DroYak_CAF1 4181 81 - 1 AACAGCAA-UGACCAGCCAAUUAAAACUGCCAAAAACUAAUGCCACCGCAAAACUAUGGUUGCUAGUUUUAUGGC------------------CG-------------CGAAA ....((..-((......)).........(((((((((((..((.((((........)))).))))))))).))))------------------.)-------------).... ( -16.90) >DroAna_CAF1 4262 105 - 1 AAAAACACUUGACCAGGCAAUUAAAACUGCCAAAAACUAAUGCCACCGCAA-ACUACGGUUUCCAGUUUUAUGGCUGCCA-------ACUGGCCAUCCCCUCUAGAUUCGAAA .(((((....((((.((((........)))).........(((....))).-.....))))....)))))((((((....-------...))))))................. ( -20.90) >consensus AACGGCAC_UGACCGGCCAAUUAAAACUGCCAAAAACUAAUGCCACCGCAAAACUACGGUUGCUAGUUUUAUGGCAGCCA_______ACUGGCCG_____________CGAAA ..........(.((((..........(((((((((((((.....((((........))))...))))))).))))))...........)))).)................... (-15.58 = -16.92 + 1.33)



| Location | 26,929,174 – 26,929,265 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -21.11 |
| Energy contribution | -21.03 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26929174 91 + 27905053 UUUGGCAGUUUUAAUUGGCCGGUCA-GCGCCGUUGGCCAA-GUUUUCAGCUGC---CCAGCGCUUCAGCUGAAAUGACAUUUCGUUAUUGUGCUGG------------- ...(((((((....(((((((((..-..)))...))))))-......))))))---)((((((...(((.(((((...))))))))...)))))).------------- ( -33.00) >DroSec_CAF1 3930 91 + 1 UUUGGCAGUUUUAAUUGGCCGGUCA-GCGCCGUUGGCCAC-GUUUUCAGCUGC---CCAGCGCUUCAGCUGAAAUGACAUUUCGUUAUUGUGCUGG------------- ...(((((((..(((((((((((..-..)))...))))).-)))...))))))---)((((((...(((.(((((...))))))))...)))))).------------- ( -32.00) >DroSim_CAF1 3982 91 + 1 UUUGGCAGUUUUAAUUGGCCGGUCA-GCGCCGUUGGCCAU-GUUUCCAGCUGC---CCAGCGCUUCAGCUGAAAUGACAUUUCGUUAUUGUGCUGG------------- ...(((((((..(((((((((((..-..)))...))))).-)))...))))))---)((((((...(((.(((((...))))))))...)))))).------------- ( -32.00) >DroEre_CAF1 4122 91 + 1 UUUGGCAGUUUUAAUUGGCCGGCCA-GUGCCGUUGGCCAC-GUUUUCAGCUGC---CCAGCGCUGCAACUGAAAUGACAUUUCGUUAUUGUGCUGG------------- ...(((((((..(((((((((((..-.....)))))))).-)))...))))))---)((((((...(((.(((((...))))))))...)))))).------------- ( -34.30) >DroYak_CAF1 4229 94 + 1 UUUGGCAGUUUUAAUUGGCUGGUCA-UUGCUGUUGGCCAC-GUUUUCAGCUGCUGGCCAAAGCUGCAACUGAAAUGACAUUUCGUUAUUGUGCUGG------------- ....(((((((....((((..((..-..((((..(((...-)))..)))).))..)))))))))))......((((((.....)))))).......------------- ( -28.40) >DroAna_CAF1 4333 107 + 1 UUUGGCAGUUUUAAUUGCCUGGUCAAGUGUUUUUGGCCAUAGUUUCUAGCCUU--GGAAUUGCAGCAGCUAAAAUAACAUUUUGUUAUUGGAUUGGUGGUGGAUGCCCA ...((((......((..((((((((((....))))))))(((((.((.((...--......)))).))))).((((((.....)))))).....))..))...)))).. ( -27.30) >consensus UUUGGCAGUUUUAAUUGGCCGGUCA_GCGCCGUUGGCCAC_GUUUUCAGCUGC___CCAGCGCUGCAGCUGAAAUGACAUUUCGUUAUUGUGCUGG_____________ ....((((((..(((((((((((.....)))...)))))..)))...))))))...(((((((...(((.(((((...))))))))...)))))))............. (-21.11 = -21.03 + -0.08)



| Location | 26,929,174 – 26,929,265 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -14.96 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
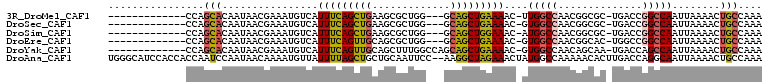
>3R_DroMel_CAF1 26929174 91 - 27905053 -------------CCAGCACAAUAACGAAAUGUCAUUUCAGCUGAAGCGCUGG---GCAGCUGAAAAC-UUGGCCAACGGCGC-UGACCGGCCAAUUAAAACUGCCAAA -------------.((((........(((((...))))).((....))))))(---((((........-((((((...((...-...))))))))......)))))... ( -25.94) >DroSec_CAF1 3930 91 - 1 -------------CCAGCACAAUAACGAAAUGUCAUUUCAGCUGAAGCGCUGG---GCAGCUGAAAAC-GUGGCCAACGGCGC-UGACCGGCCAAUUAAAACUGCCAAA -------------.((((........(((((...))))).((....))))))(---((((........-.(((((...((...-...))))))).......)))))... ( -25.99) >DroSim_CAF1 3982 91 - 1 -------------CCAGCACAAUAACGAAAUGUCAUUUCAGCUGAAGCGCUGG---GCAGCUGGAAAC-AUGGCCAACGGCGC-UGACCGGCCAAUUAAAACUGCCAAA -------------.((((........(((((...))))).((....))))))(---((((.((....)-)(((((...((...-...))))))).......)))))... ( -28.60) >DroEre_CAF1 4122 91 - 1 -------------CCAGCACAAUAACGAAAUGUCAUUUCAGUUGCAGCGCUGG---GCAGCUGAAAAC-GUGGCCAACGGCAC-UGGCCGGCCAAUUAAAACUGCCAAA -------------.((((.(..(((((((((...))))).))))..).))))(---((((........-.(((((...(((..-..)))))))).......)))))... ( -29.99) >DroYak_CAF1 4229 94 - 1 -------------CCAGCACAAUAACGAAAUGUCAUUUCAGUUGCAGCUUUGGCCAGCAGCUGAAAAC-GUGGCCAACAGCAA-UGACCAGCCAAUUAAAACUGCCAAA -------------...(((..........((((..((((((((((.((....))..))))))))))))-))(((...((....-))....))).........))).... ( -23.00) >DroAna_CAF1 4333 107 - 1 UGGGCAUCCACCACCAAUCCAAUAACAAAAUGUUAUUUUAGCUGCUGCAAUUCC--AAGGCUAGAAACUAUGGCCAAAAACACUUGACCAGGCAAUUAAAACUGCCAAA (((.(((...............((((.....))))((((((((..((......)--).))))))))...))).)))..............((((........))))... ( -20.40) >consensus _____________CCAGCACAAUAACGAAAUGUCAUUUCAGCUGAAGCGCUGG___GCAGCUGAAAAC_GUGGCCAACGGCAC_UGACCGGCCAAUUAAAACUGCCAAA ................(((................(((((((((.............)))))))))....(((((..............)))))........))).... (-14.96 = -14.96 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:47:46 2006