| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,920,631 – 26,920,736 |
| Length | 105 |
| Max. P | 0.551667 |

| Location | 26,920,631 – 26,920,736 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -10.74 |
| Energy contribution | -10.88 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
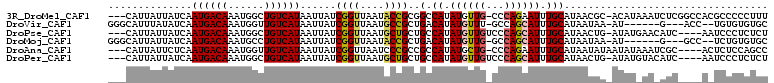
>3R_DroMel_CAF1 26920631 105 + 27905053 ---CAUUAUUAUCAAUGACAAAUGGCUGUCAUAAUUAUCGGUUAAUACCGCGGCCAUAUGUUG-CCCAGAAUUUGCAUAACGC-ACAUAAAUCUCGGCCACGCCCCCUUU ---.............((((.(((((((..........((((....))))))))))).))))(-((.(((((.(((.....))-).))...))).)))............ ( -23.40) >DroVir_CAF1 39193 97 + 1 GGGCAUUUAUAUCAAUGACAAAUGGUUGUCAUAAUUAUCGGUUAAUGCCGCUGACAUAUGUUU-GCCAGCAUUUGCAUAAUAA-AU------G---ACC--UGUGUGUGC .((((..(((((..(((((((....)))))))..(((.((((....)))).))).)))))..)-))).((((..(((((....-..------.---...--))))))))) ( -24.90) >DroPse_CAF1 38747 102 + 1 ---CAUUAUUAUCAAUGACAAAUGGCUGUCAUAAUUAUCGGUUAAUGCUGCUGCCAUAUGUUGUCCCAGCAUUUGCAUAACUG-AUAUGAACAUC----AAUCCCUCUCU ---........(((((((((......))))))...(((((((((.((((((((.((.....))...)))))...)))))))))-)))))).....----........... ( -23.30) >DroMoj_CAF1 42330 97 + 1 GGGCAUUAUUAUCAAUGACAAAUGCCUGUCAUAAUUAUCGGUUAAUACCGCUGACAUAUGUUG-GCCAGCAUUUGCAUAAUAA-AU------G---GCC--UCUGUGUGC ((((.((((((....((.(((((((.((...........(((....)))((..((....))..-)))))))))))))))))))-..------.---)))--)........ ( -25.00) >DroAna_CAF1 29085 102 + 1 ---CAUUAUUCUCAAUGACAAAUGGUUGUCAUAAUUAUCGGUUAAUCCCGCCGCCAUAUGCUG-CCCAGAAUUUGCAUAAUAUAAUAUAAAUCGC----ACUCUCCAGCC ---...........(((((((....)))))))......((((.......))))......((((-...(((...(((.................))----).))).)))). ( -14.23) >DroPer_CAF1 38595 102 + 1 ---CAUUAUUAUCAAUGACAAAUGGCUGUCAUAAUUAUCGGUUAAUGCUGCUGCCAUAUGUUGUCCCAGCAUUUGCAUAACUG-AUAUGUACAUC----AAUCCCUCUCU ---...........((((((......))))))...(((((((((.((((((((.((.....))...)))))...)))))))))-)))........----........... ( -22.20) >consensus ___CAUUAUUAUCAAUGACAAAUGGCUGUCAUAAUUAUCGGUUAAUGCCGCUGCCAUAUGUUG_CCCAGCAUUUGCAUAACAA_AUAU_AA_AUC____A_UCCCUCUCC ..............((((((......))))))......((((....))))..(.((.((((((...)))))).))).................................. (-10.74 = -10.88 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:47:39 2006